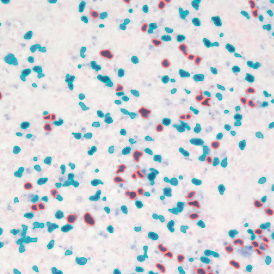
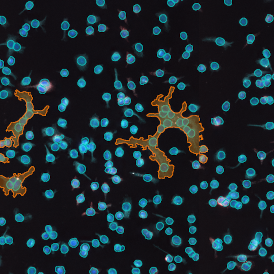
Leishmaniasis parasite detection in cell culture and tissue: Leishmaniasis APP
The Leishmaniasis APP within TissueGnostics contextual image analysis software, StrataQuest, was designed to assist in immunology research into Leishmaniasis and can be adapted for research on other intracellular parasites. It can be used both on cell cultures and tissue sections. Its main function is to automatically detect parasite stages in cell compartments and quantify them in context with specific immune markers. In this case study, we will show in detail how simple and straightforward the analysis is.
The aim of this project was to detect how many cells are infected by leishmaniasis and the number of leishmaniasis parasites per cell.
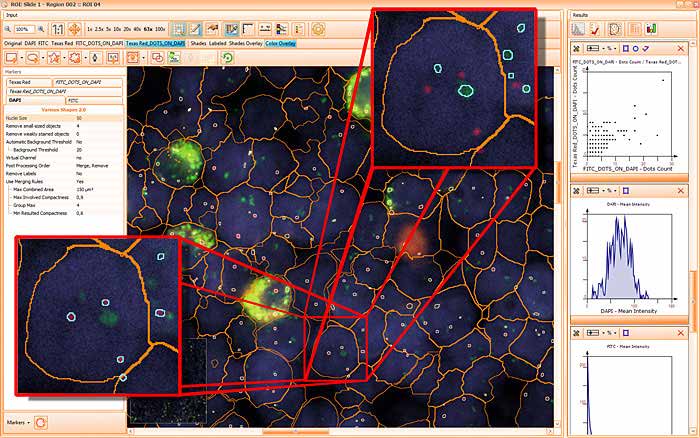
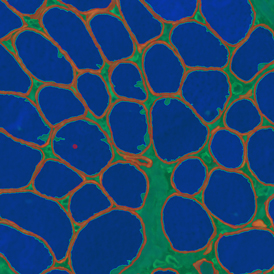
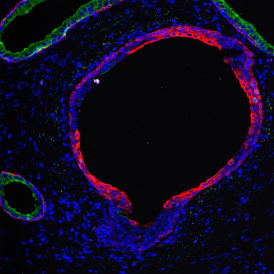
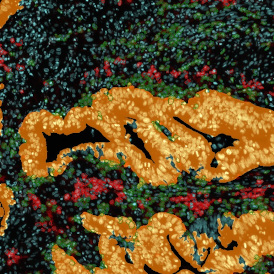
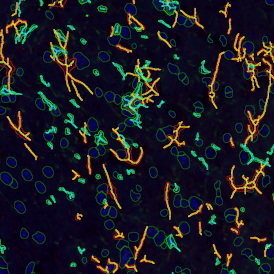
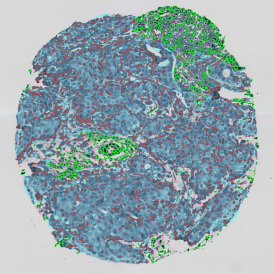
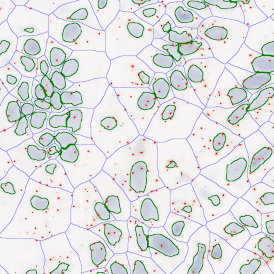
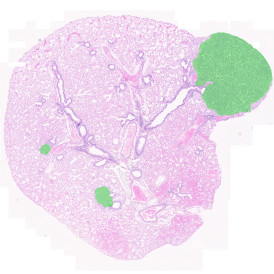
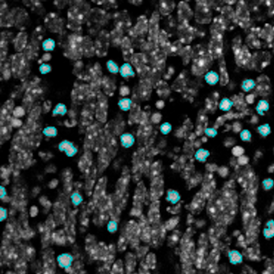
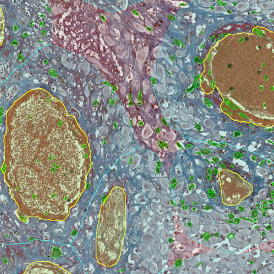
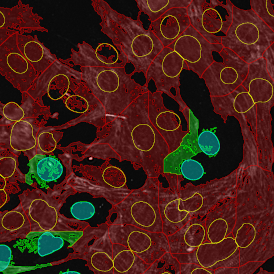
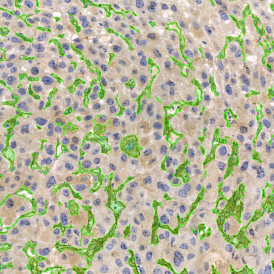
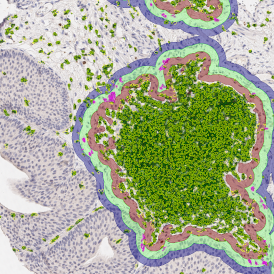
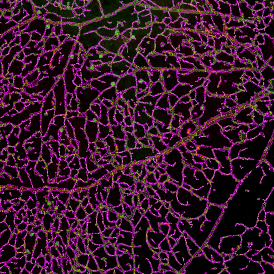
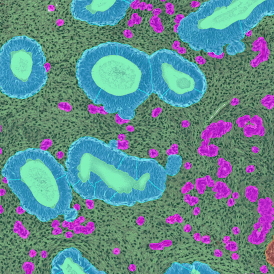
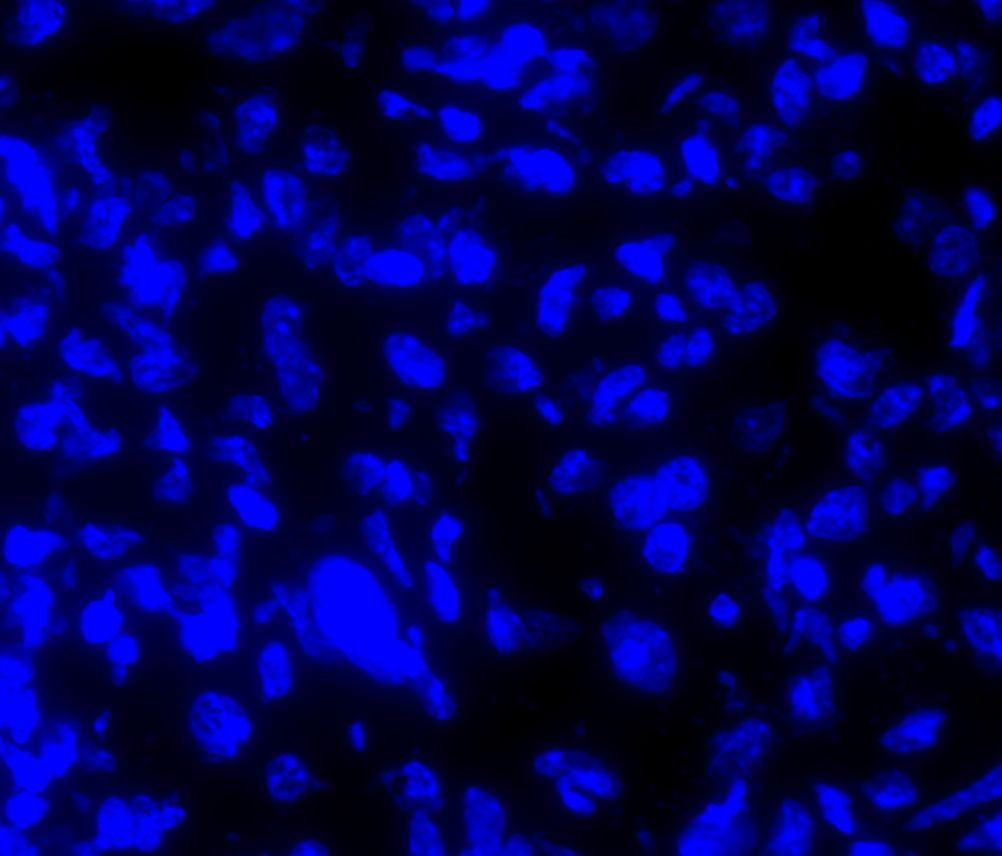
The image below shows a mouse skin-draining lymph node stained with DAPI (nucleus, blue).
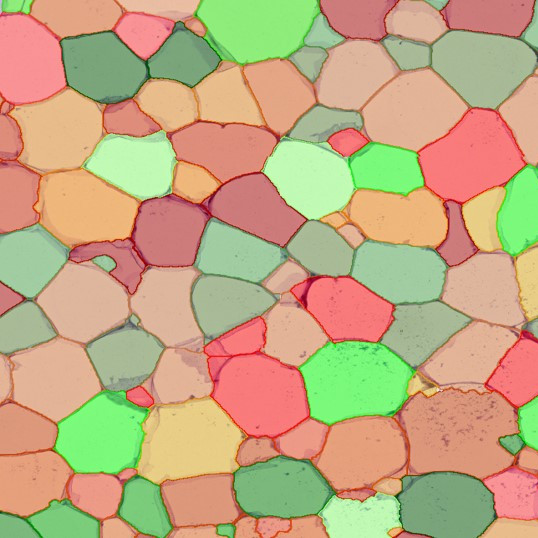
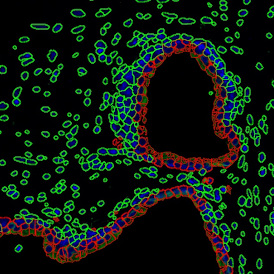
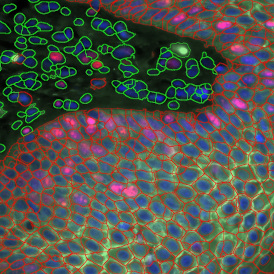
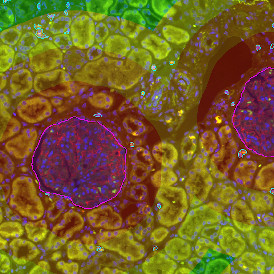
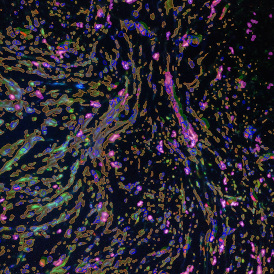
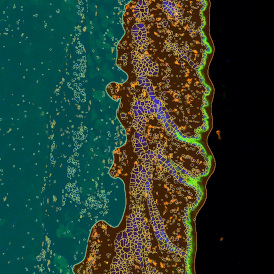
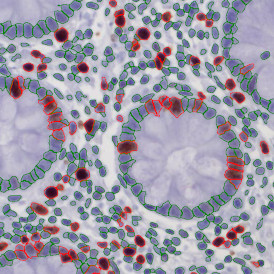
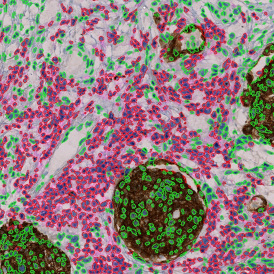
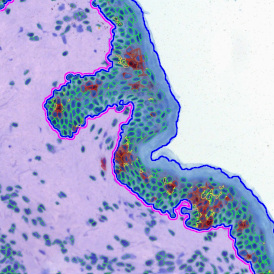
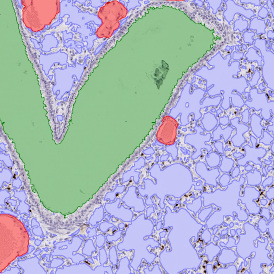
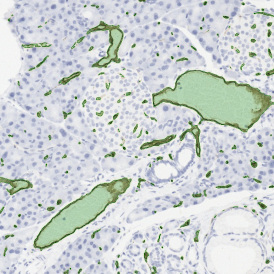
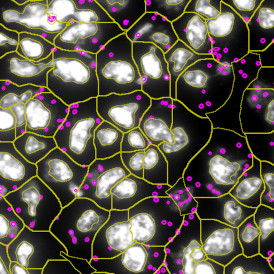
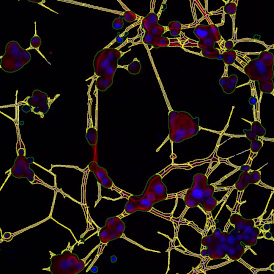
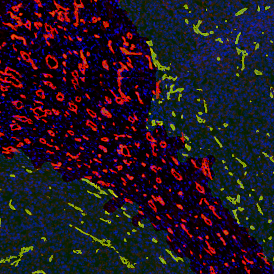
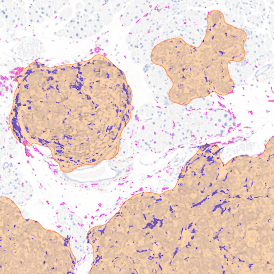
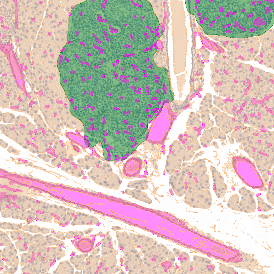
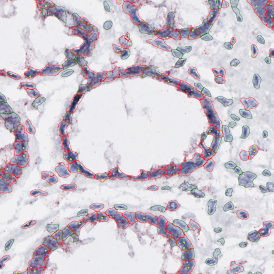
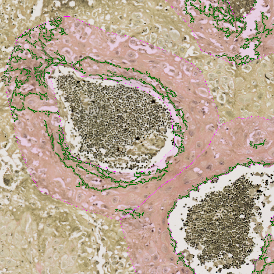
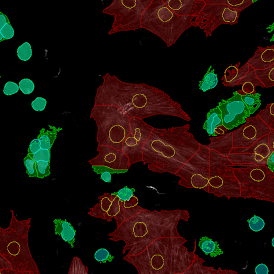
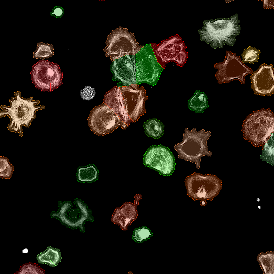
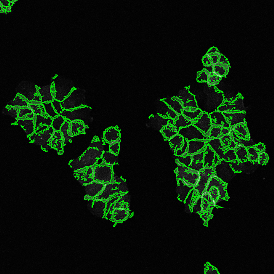
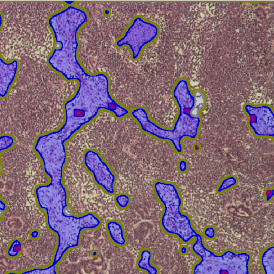
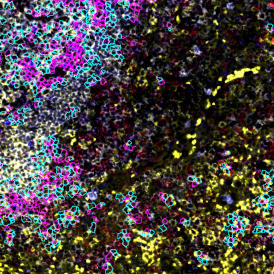
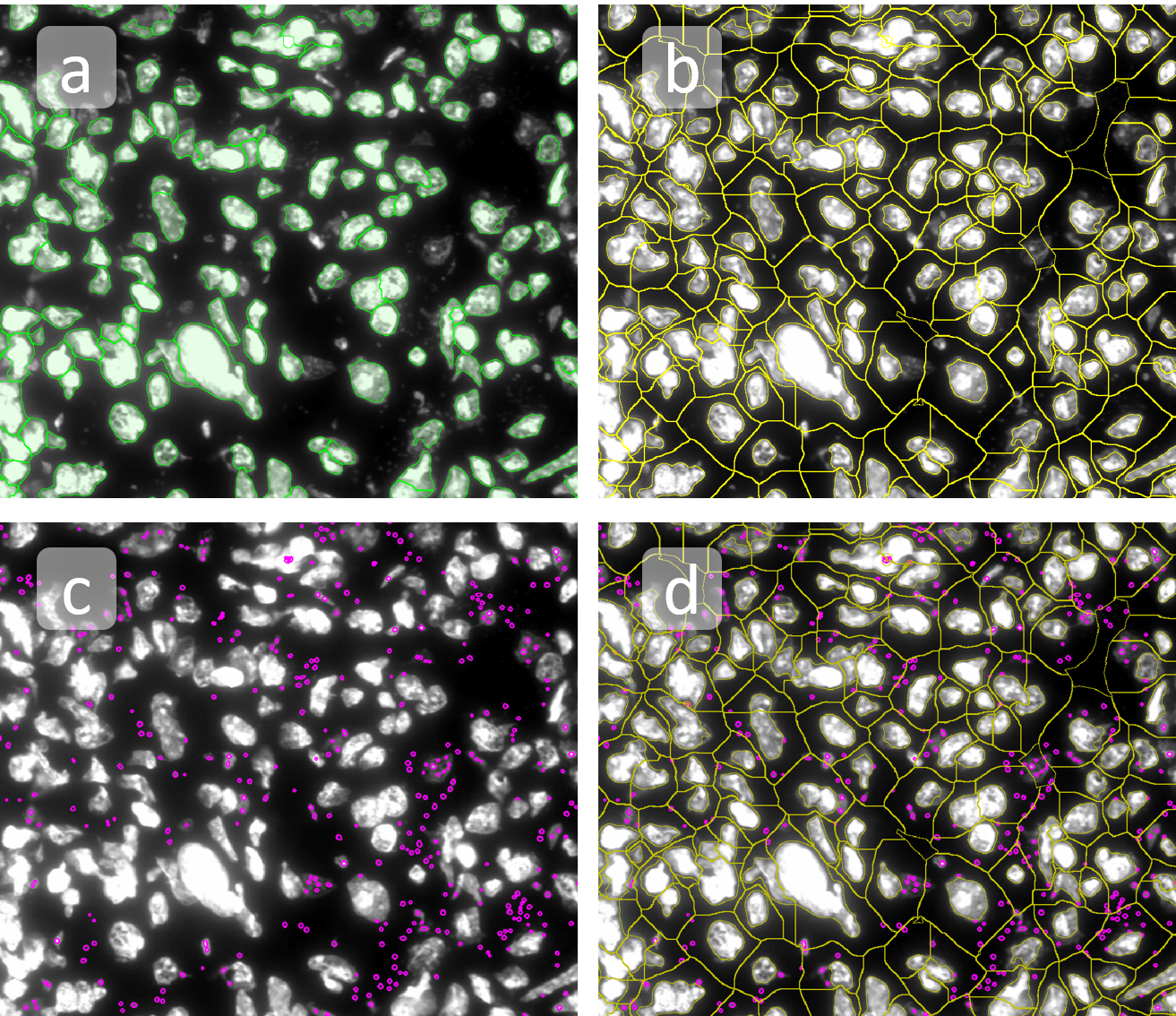
Nuclei detection based on DAPI staining is one of the most crucial first steps in image analysis; (a) here nuclei are outlined in green. Starting from this, the software defines the whole cell around the nuclei, outlined in yellow for representation (b); (c) detection of parasites inside the cell as shown here as pink dots; (d) combined masks showing cells in yellow and Leishmaniasis parasites in pink.
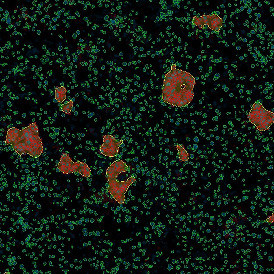
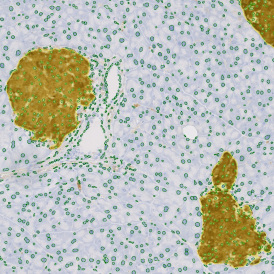
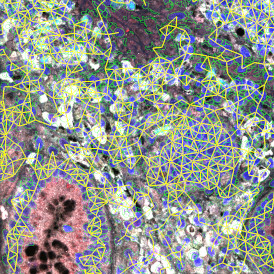
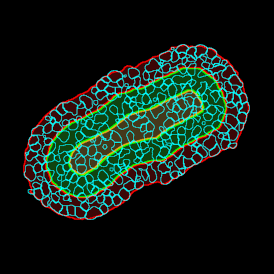
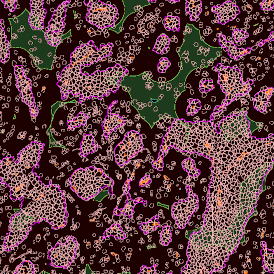
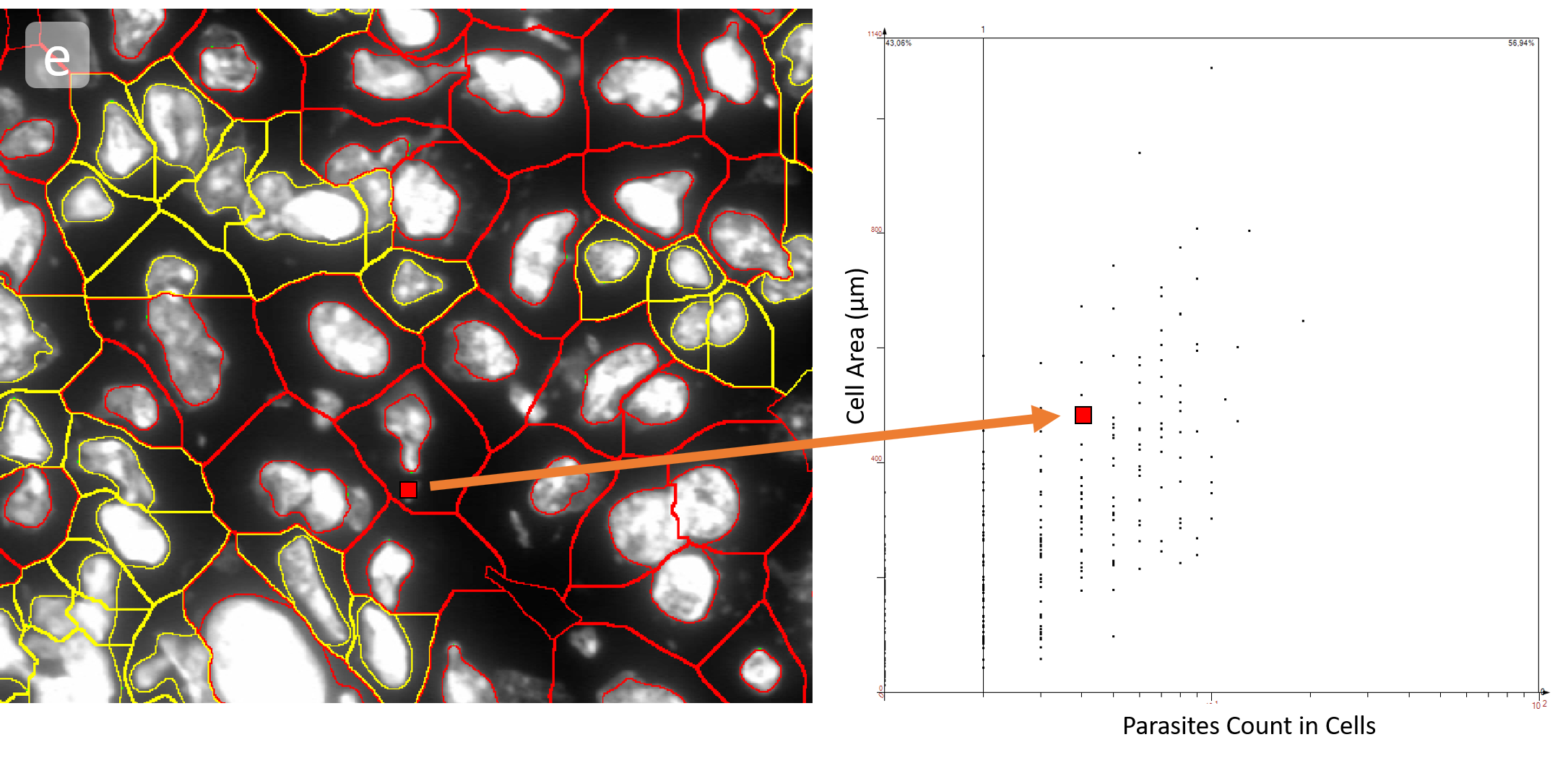
The obtained data is visualized within StrataQuest using scattergrams. In the example below, (e) cells containing parasites are highlighted in red by selecting them in the proper scattergram (in this case, parasites count vs cell size). The individual cells within an image can be chosen and their exact position in the corresponding scattergram is shown – this is a feature available in StrataQuest called forward connection, especially useful for validation of the selected gates.
In the end, all results with many parameters to choose from can be outputted into common file formats (Excel, PDF, CSV) for further statistical analysis.
However, the power of StrataQuest´s App-based streamlined workflow does not end with cell/parasite detection. For further deciphering of Leishmania life cycle aspects and distribution processes, the parasite load and the status of parasite stages, e.g. “live/dead” can also be evaluated to obtain additive information. The IF Dots APP enables the analysis of FISH-stained leishmaniasis parasites.
A recent paper utilizing StrataQuest´s analysis software showed that programmed death-ligand 1 (PD-L1) expression is predictive of clinical response to treatment in patients with leishmaniasis [2]. If you are interested in performing detailed context-based quantitative analysis, explore the potential of StrataQuest and StrataQuest Apps.
Sources:
- Doehl JSP, Ashwin H, Brown N, Romano A, Carmichael S, Pitchford JW, Kaye PM. Spatial Point Pattern Analysis Identifies Mechanisms Shaping the Skin Parasite Landscape in Leishmania donovani Infection. Front Immunol. 2021 Dec 16;12:795554. doi: 10.3389/fimmu.2021.795554.
- Nidhi S. Dey, Paul M. Kaye, Shalindra Ranasinghe. Early reduction in PD-L1 expression predicts faster treatment response in human cutaneous leishmaniasis. October 5, 2021. J Clin Invest. 2021;131(22):e142765. doi.org/10.1172/JCI142765.
- TissueGnostics, 2022. White paper. PD-L1 raises new hope in the treatment of cutaneous leishmaniasis.

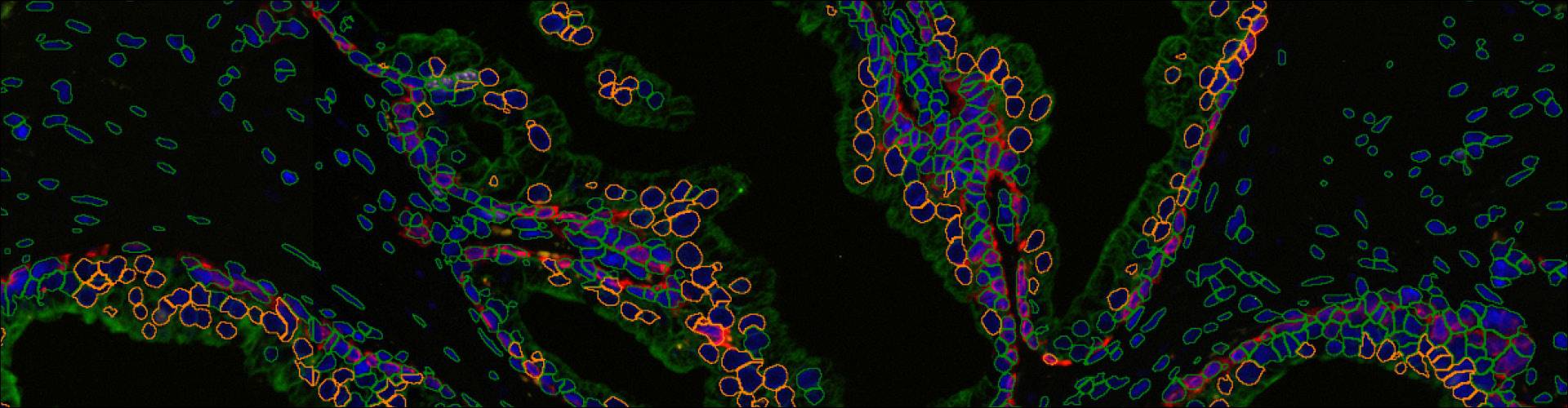

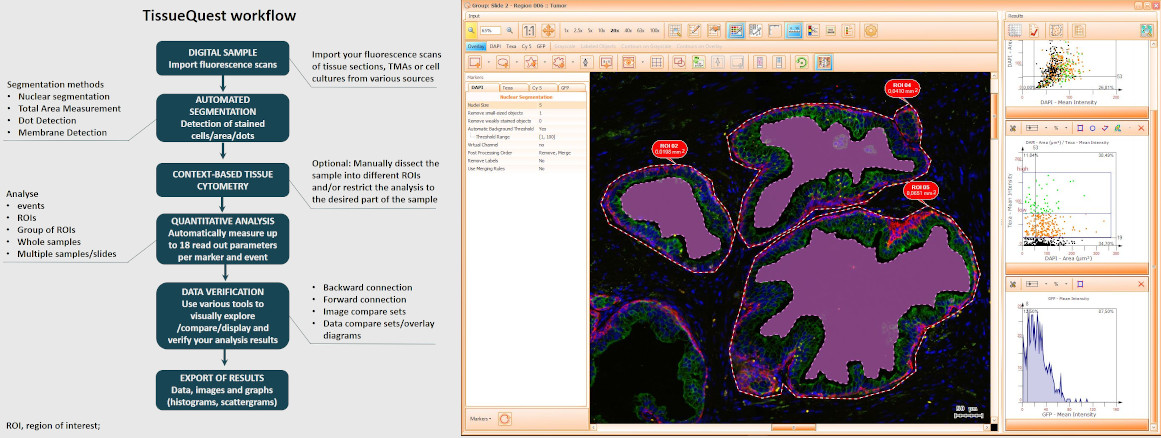
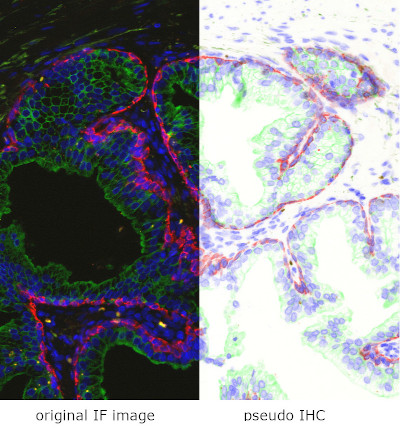
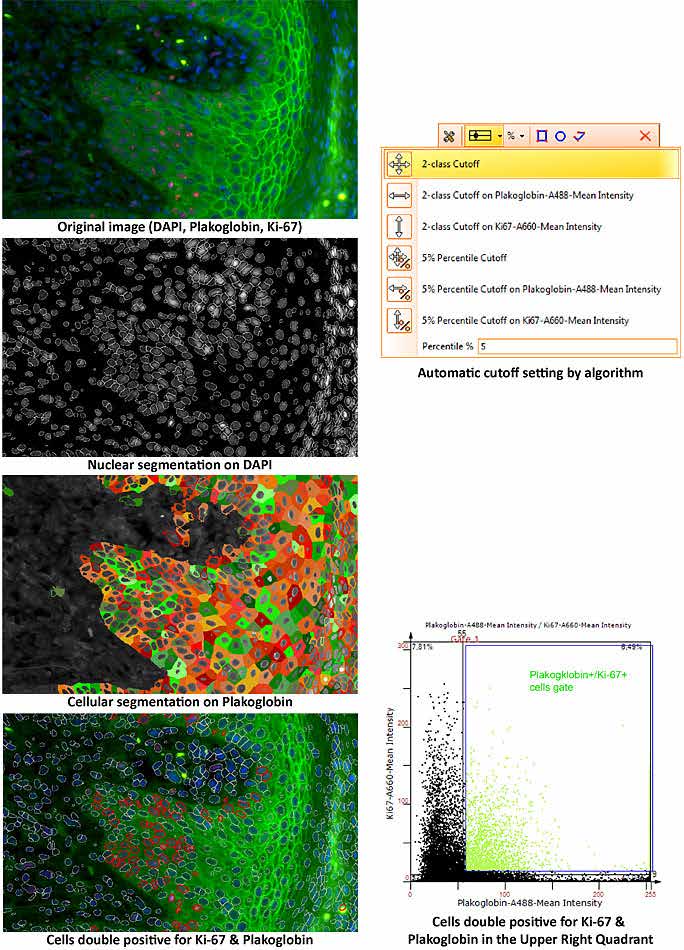 TissueQuest uses algorithms for nuclear and cell compartment segmentation, algorithms for stained area detection as well as algorithms for the detection of dots inside already detected cell compartments.
TissueQuest uses algorithms for nuclear and cell compartment segmentation, algorithms for stained area detection as well as algorithms for the detection of dots inside already detected cell compartments.