TMA Analysis with StrataQuest
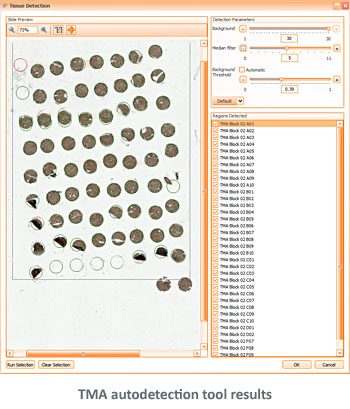
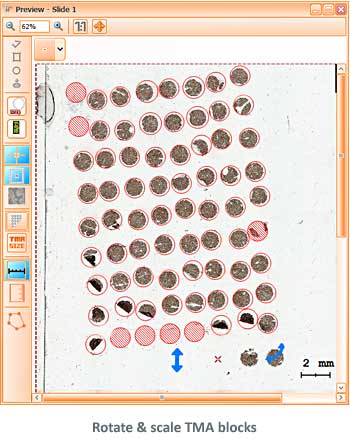
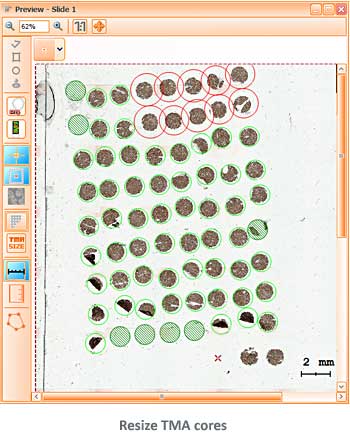
Tissue microarrays (TMAs) have proven themselves as a standard molecular clinical diagnosis technique. TMAs are paraffin blocks in which up to 1000 individual tissue cores assembled in array fashion, allowing high-throughput histological analysis.
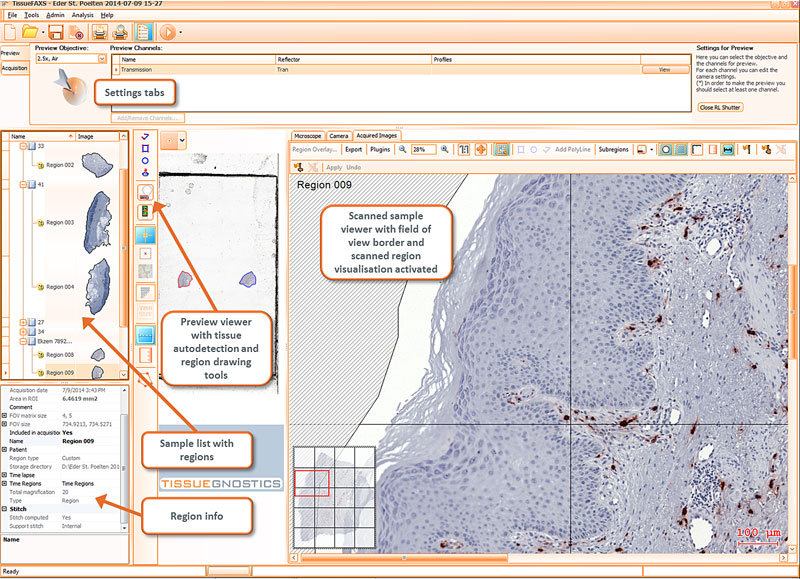
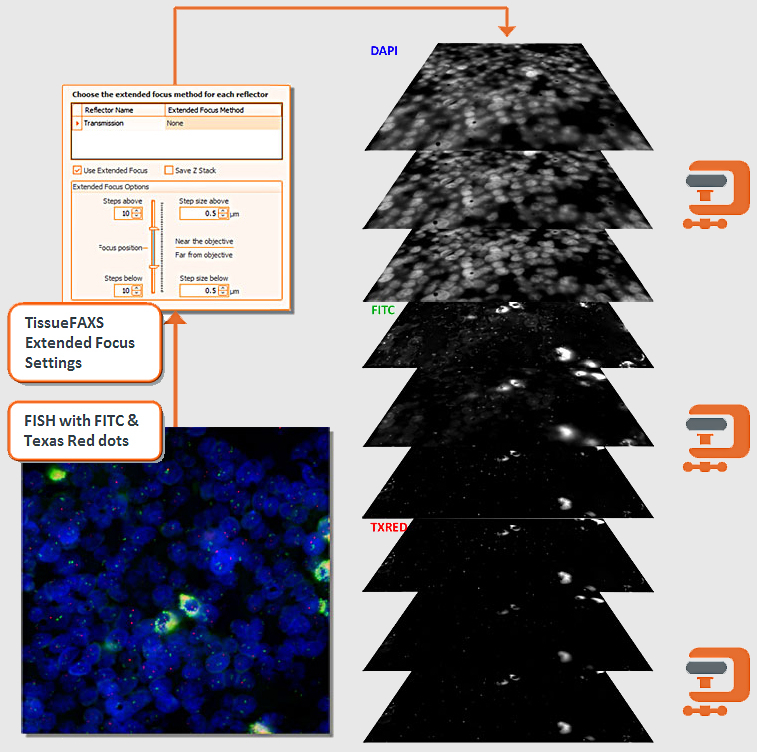
The TissueFAXS platform including the high-end image analysis solution StrataQuest provides specific workflow to scan and analyze the TMA samples containing multiple regions on one slide. TMA analysis in StrataQuest is available for images acquired in all imaging modalities (multispectral, fluorescence, brightfield).
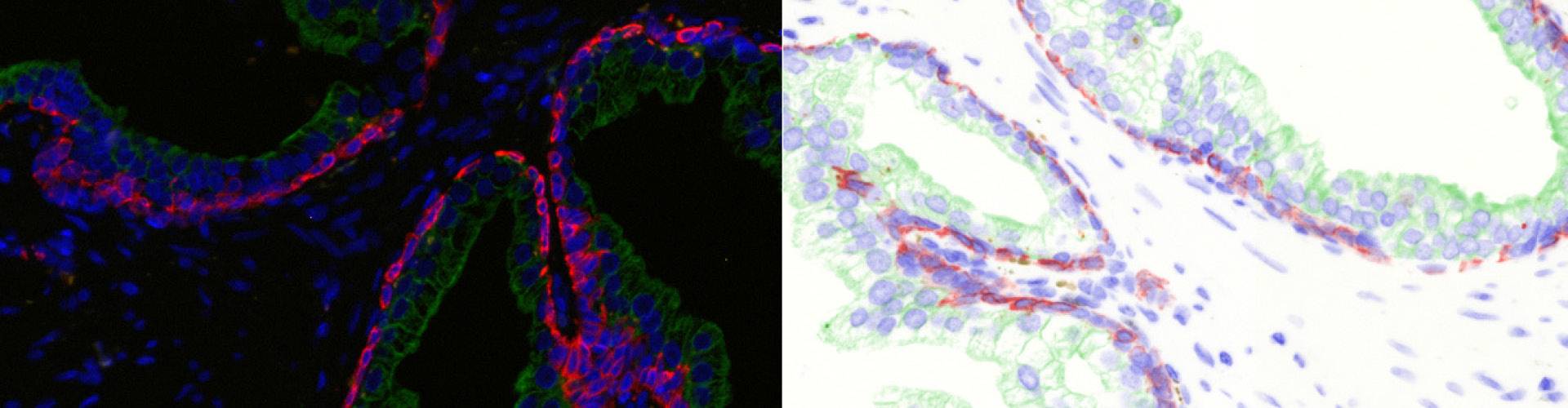
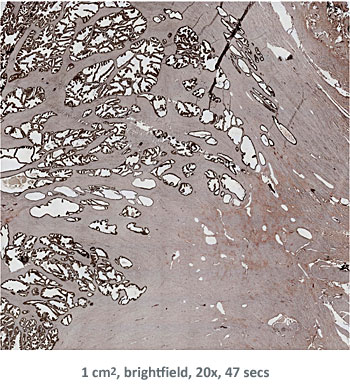
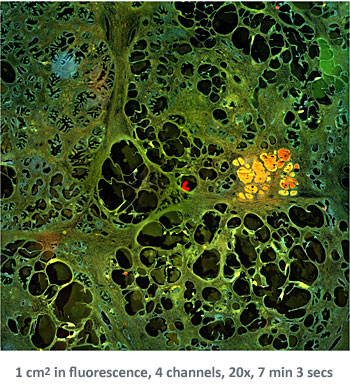
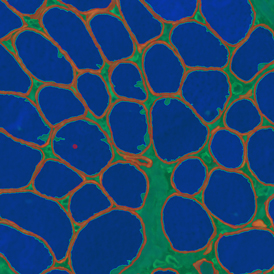
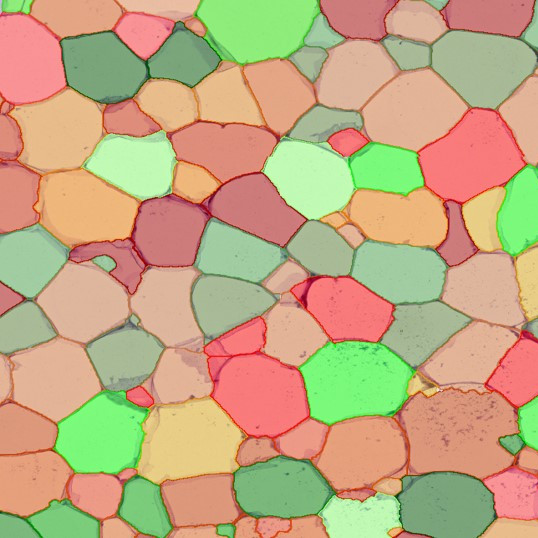
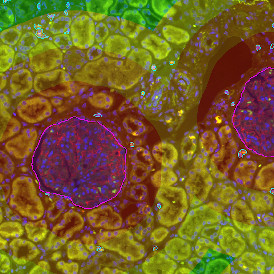
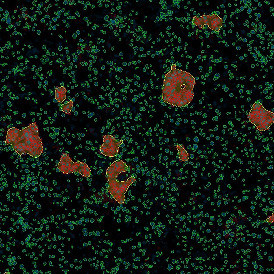
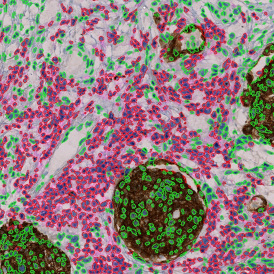
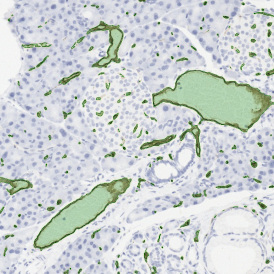
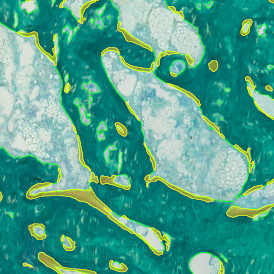
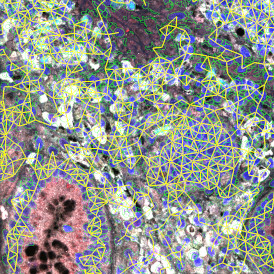
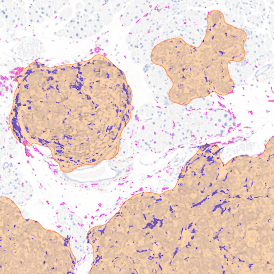
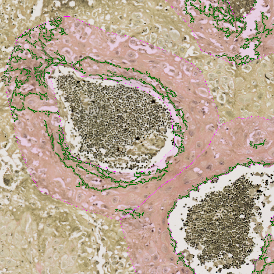
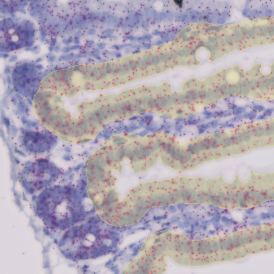
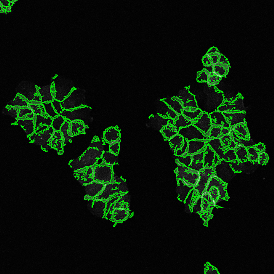
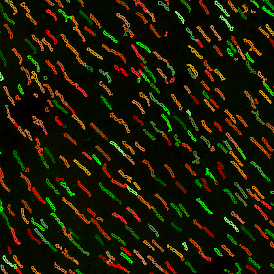
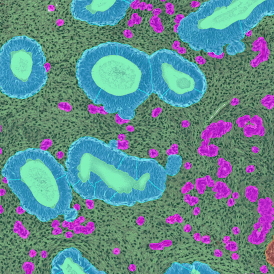
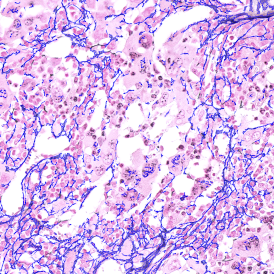
In the first example, StrataQuest was utilized to analyze a TMA sample stained for hematoxylin (blue) and for E-cadherin present in the cell membranes of epithelial cells (brown). On the original image (a), E-cadherin is shown in brown and nuclei in blue. The aim was to detect the nuclei, the epithelial cells and the membranes of epithelial cells. The challenge with this analysis was that some cells do not contain nuclei.
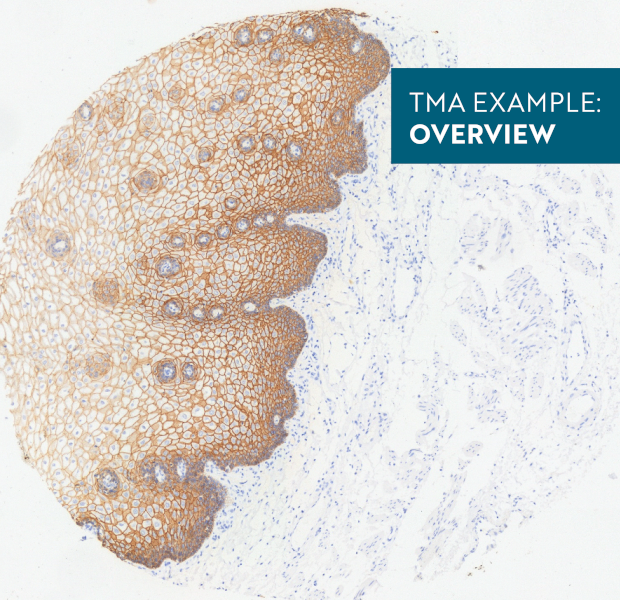
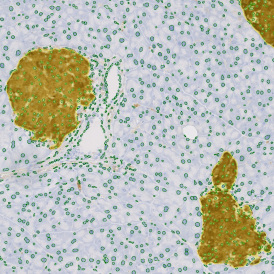
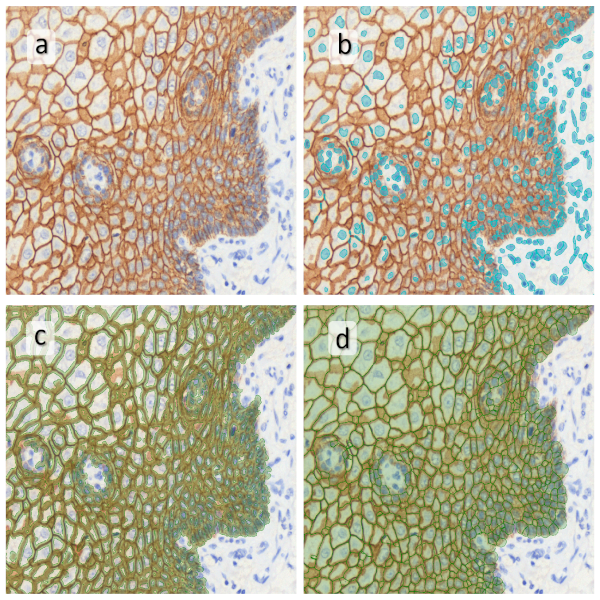
Image (b) shows detected nuclei, covered by nuclei mask in light blue. Next only membranes were detected (c), which are shown in green; (d) shows the detection of the whole E-cadherin+ epithelial tissue, also marked in green.
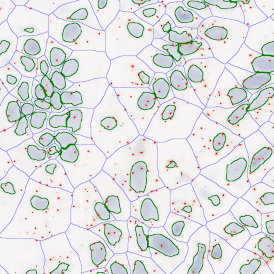
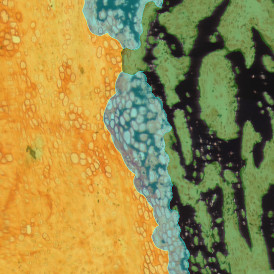
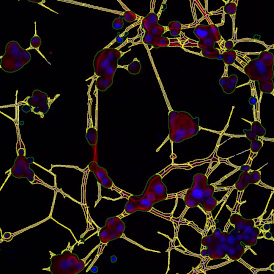
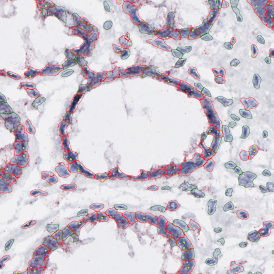
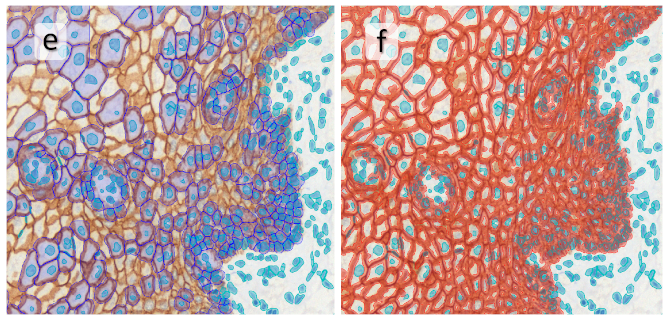
In image (e) only cells with nuclei are identified, and the whole cell with the membrane is shown in blue, while the nuclei are covered by the light blue nuclei mask; (f) represents the nuclei in light blue and the total membranes in red.
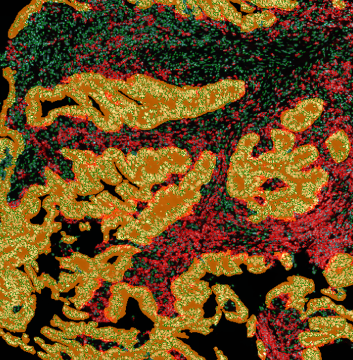
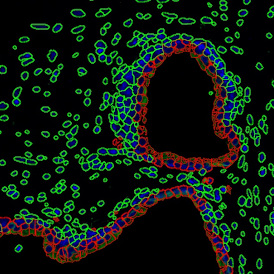
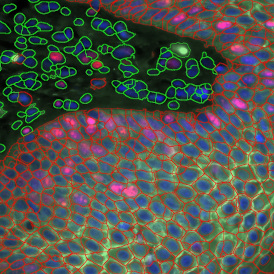
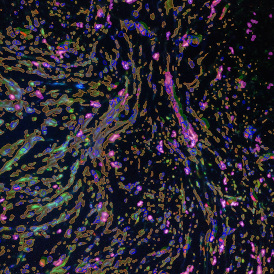
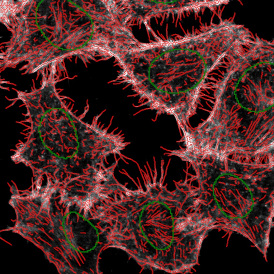
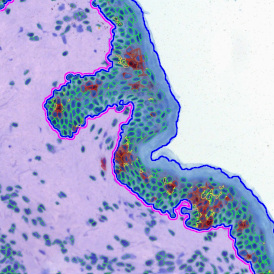
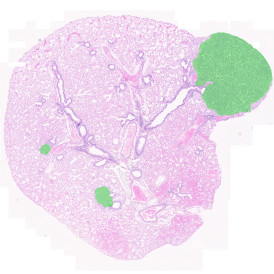
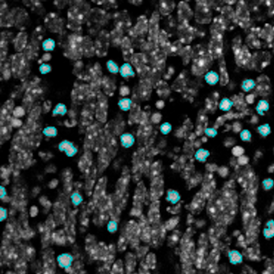
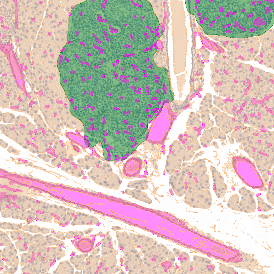
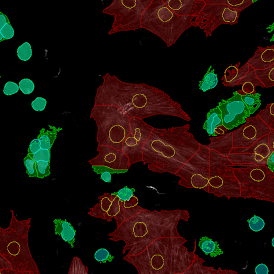
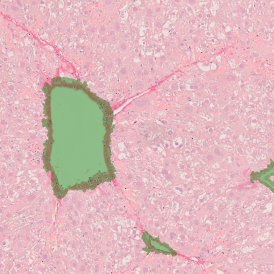
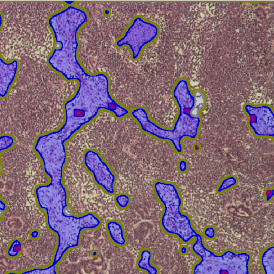
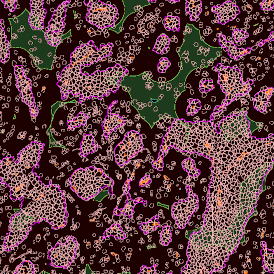
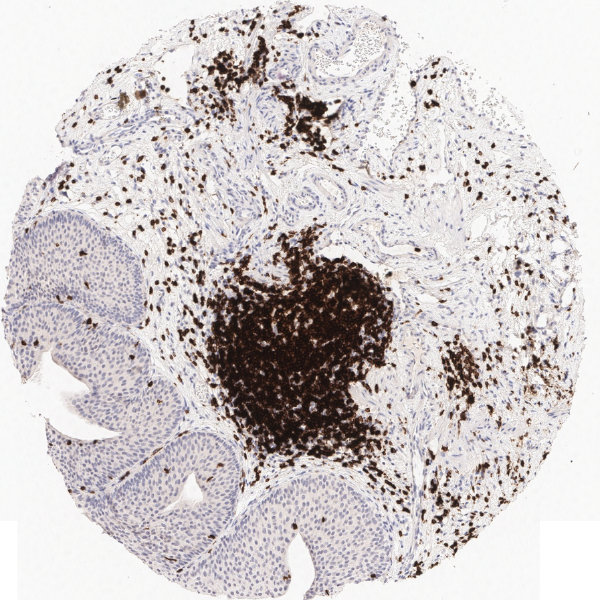
Another example on TMA analysis is our IHC Immune Status in situ APP. Here the task was to develop a streamlined workflow for tissue classification (epithelium, stroma, lymphoid cluster).
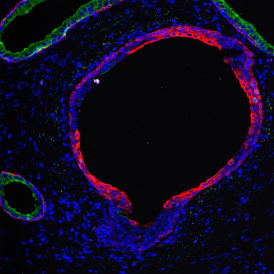
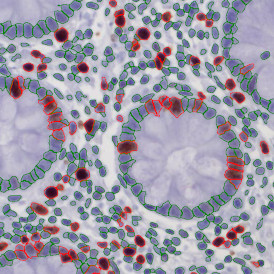
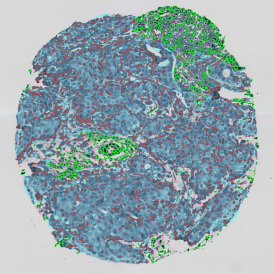
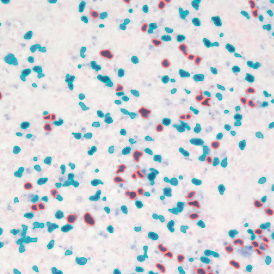
Marker positive cells (for CD3+) were identified and spatial analysis was peformed, which would allow for the calculation of how many and which types of cells are located in defined distances from the object of interest (epithelium or lymphoid cluster). In the below example the App was applied to Tissue Microarrays (TMA) of an inflamed bladder.
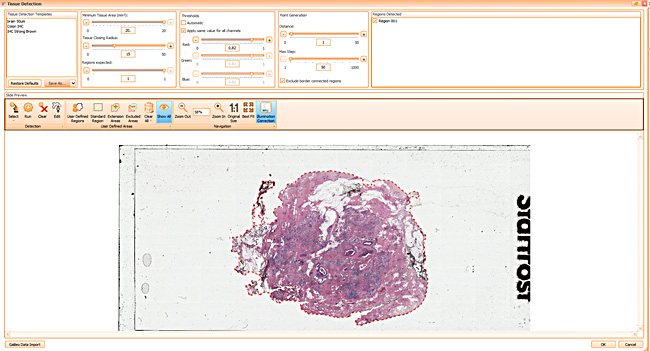
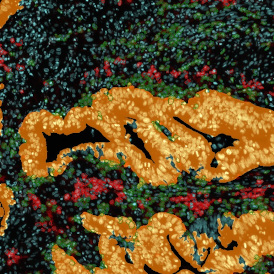
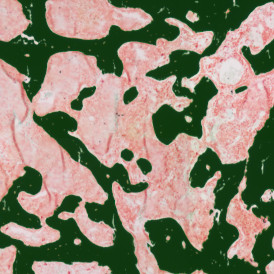
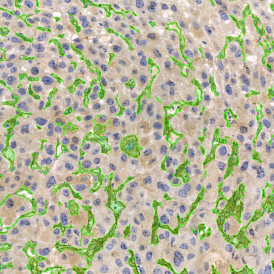
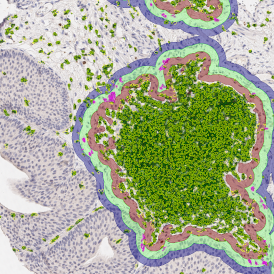
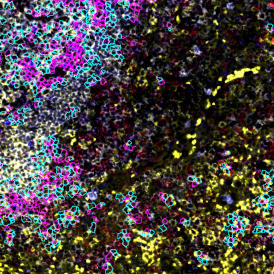
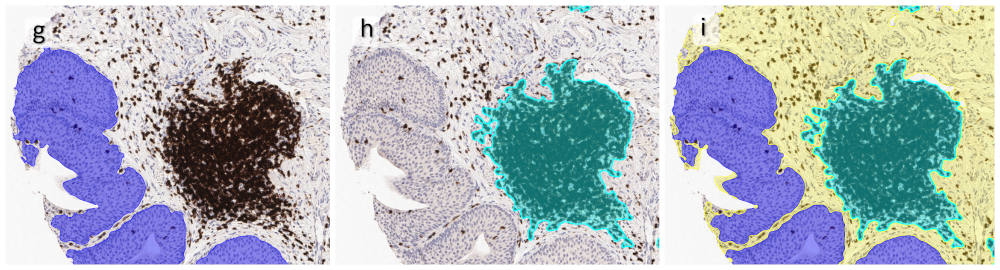
One of the most notable features of StrataQuest is the machine learning-based tissue classification. To train the algorithm, the user marks regions assigned to different tissues and trains the classifier on the sample; later this workflow can be used on other samples. The following images represent the outcome of the classifier: (g) epithelium is detected in blue; (h) displays the lymphocyte cluster in light blue. These algorithms (or masks) can be combined to achieve a better visualization: (i) epithelium is displayed in blue, lymphocyte cluster in light blue and stroma in yellow.
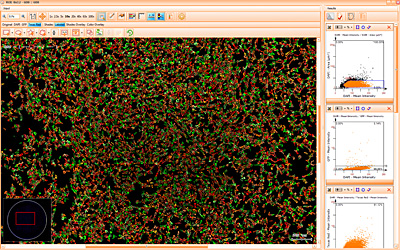
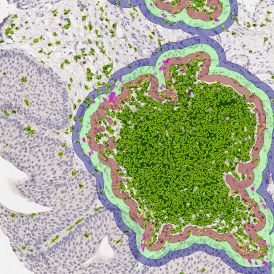
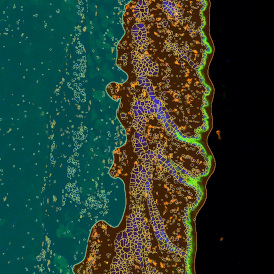
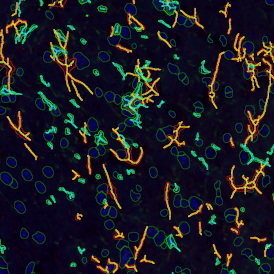
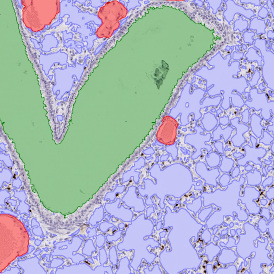
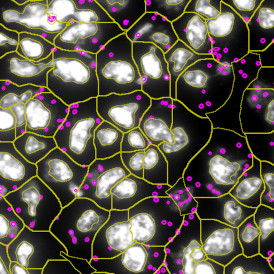
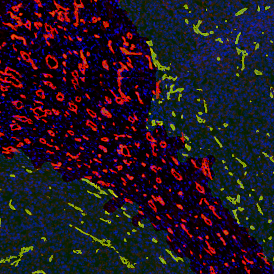
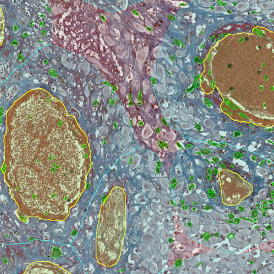
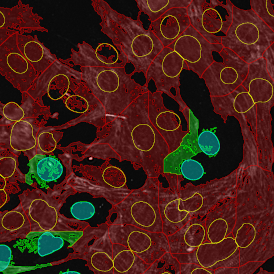
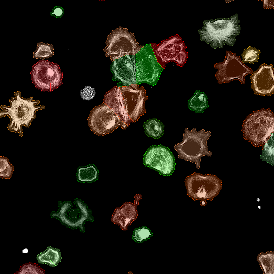
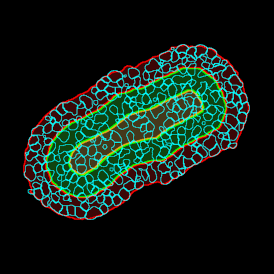
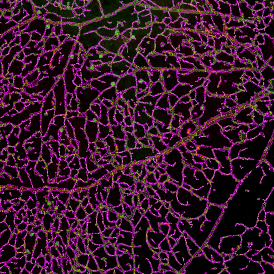
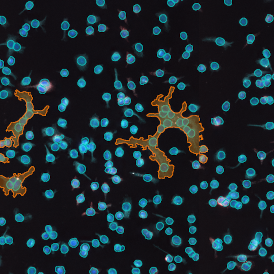
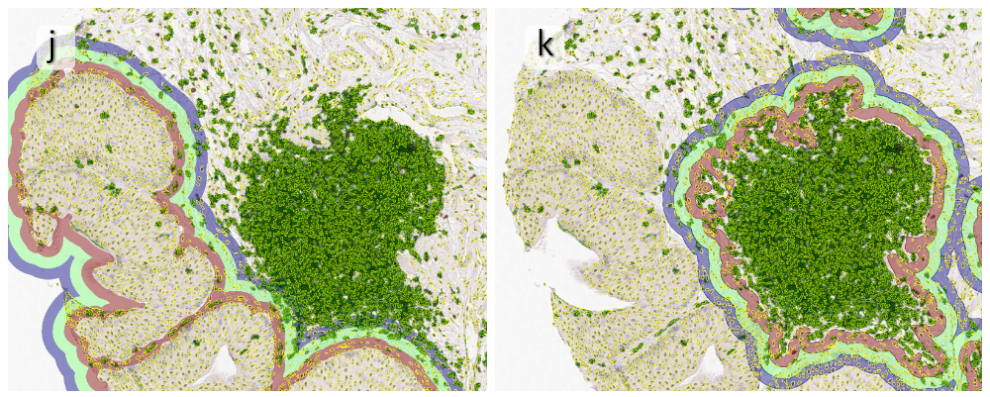
Beside the tissue classification itself, StrataQuest is capable of performing proximity analysis. It is possible to define specific proximity ranges; in the following case each range marked by different colors is 20µm. Nuclei detection was applied and CD3+ cells are shown in green, negative cells in yellow; (j) proximity ranges for the epithelium and (k) proximity ranges for the lymphocyte cluster.
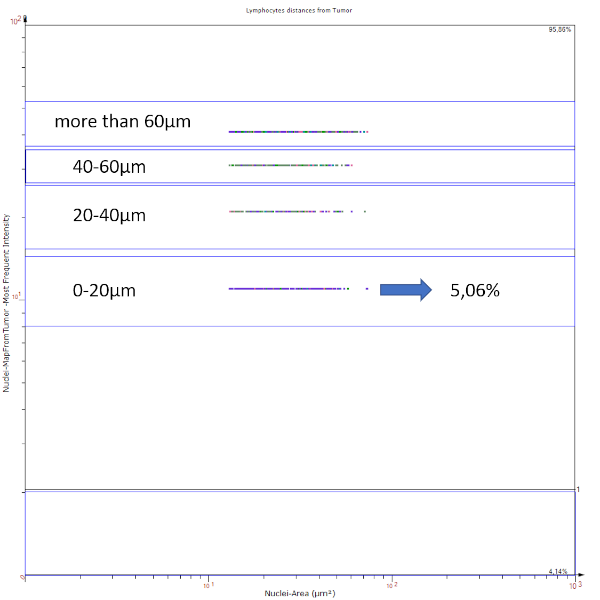
Apart from visual image analysis, StrataQuest outputs quantitative numbers. This way, it is possible to acquire and validate much more information. For example, StrataQuest can show how many cells are detected in each range: through the establishment of gates, 5.06% of CD3+ cells were found to be very close (in 20 µm range) to the epithelium.
Further details about our various products and services can be found on our website. If you are interested in learning more about TissueGnostics solutions for TMA application or other tissue cytometry applications and our related services, please reach out to a member of the TissueGnostics team.