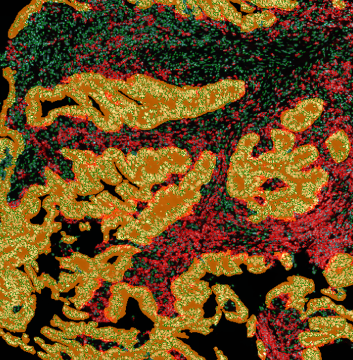
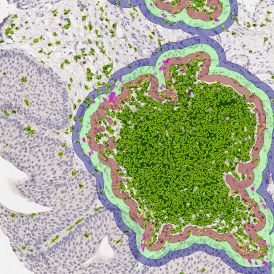
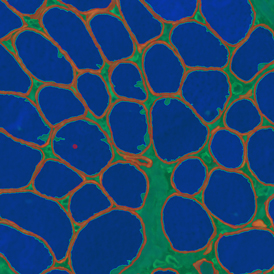
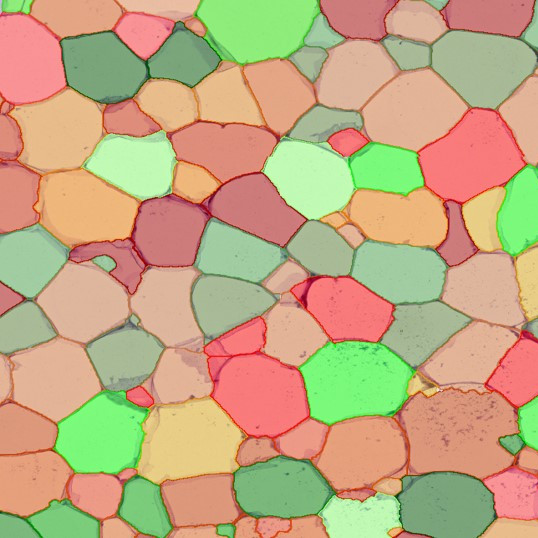
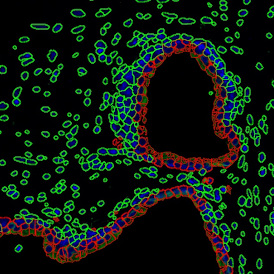
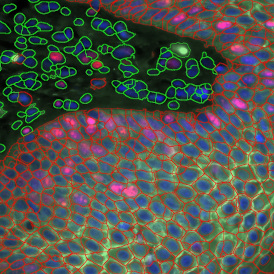
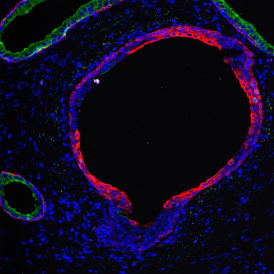
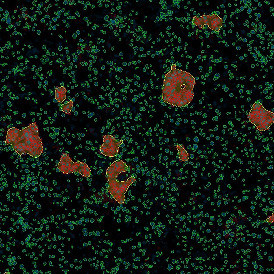
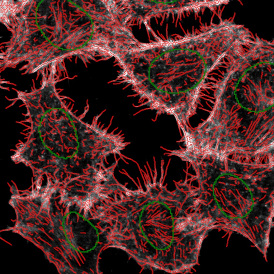
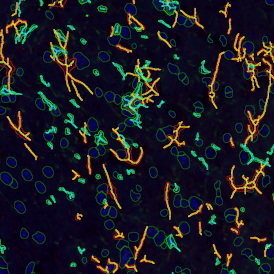
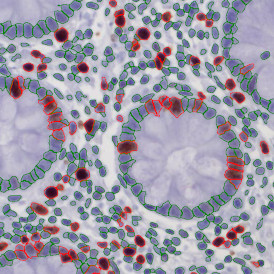
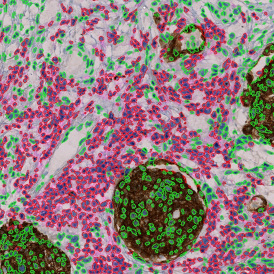
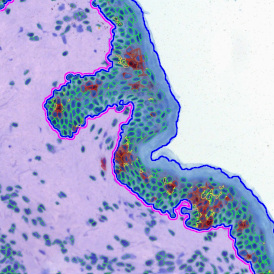
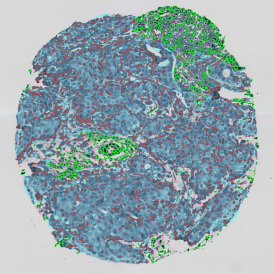
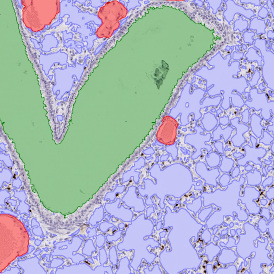
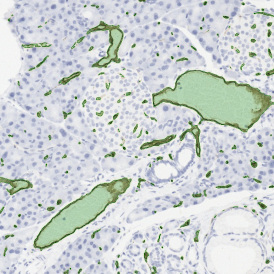
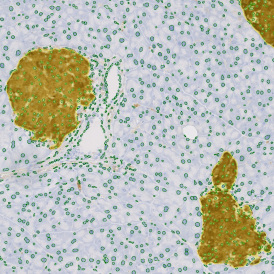
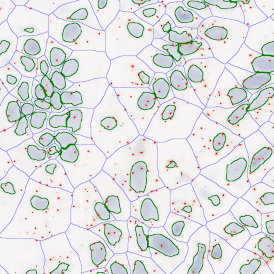
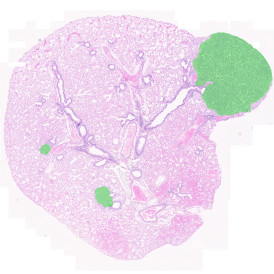
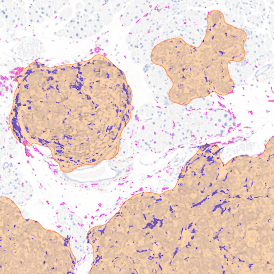
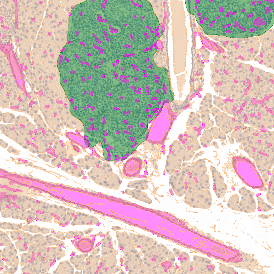
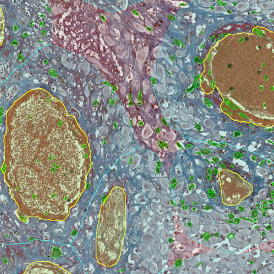
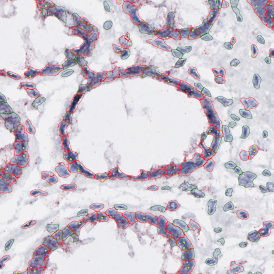
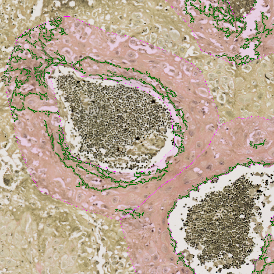
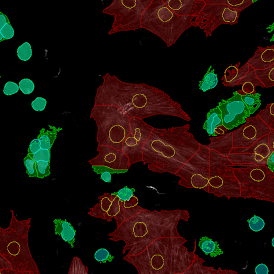
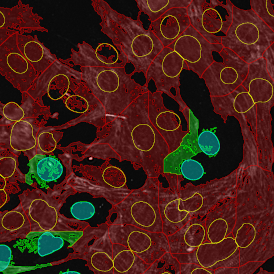
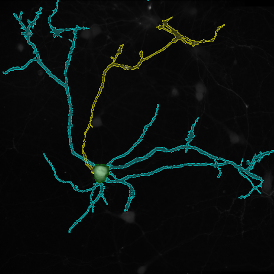
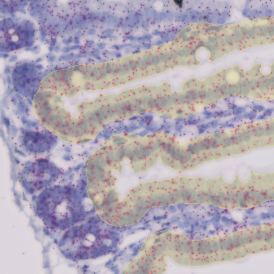
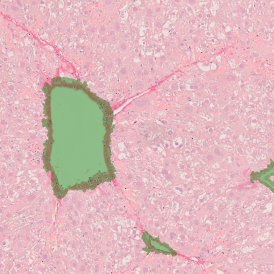
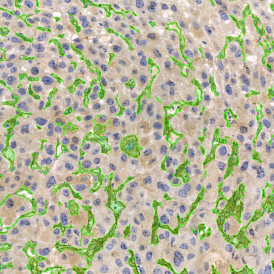
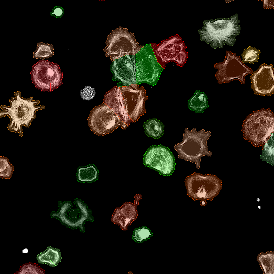
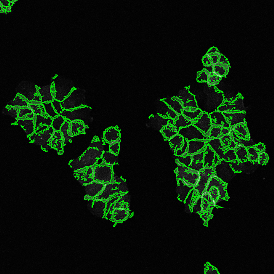
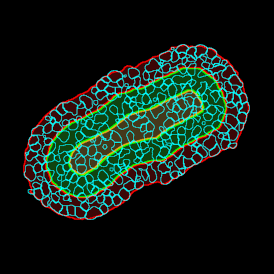
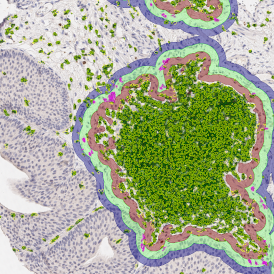
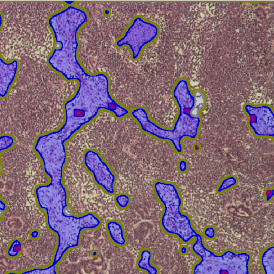
Brightfield APPs
Please follow the links to explore our StrataQuest AI solutions and Fluorescence Apps.
Contact
TissueGnostics GmbH
Taborstraße 10/2/8
1020 Vienna, Austria
+43 1 216 11 90
This email address is being protected from spambots. You need JavaScript enabled to view it.