23
OCT
23. October 2023
Understanding the Challenges of Immunofluorescence Image Analysis
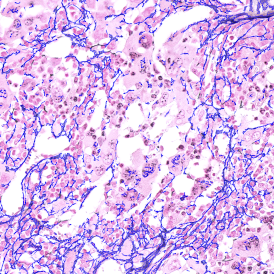
Tissue samples are highly complex in their structure. This complexity makes screening samples for signs of immune modulation as a result of disease and for understanding new therapeutic pathways a very challenging task, but one that offers a promising route to developing new therapies.1
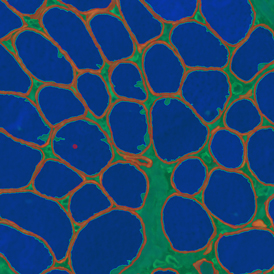
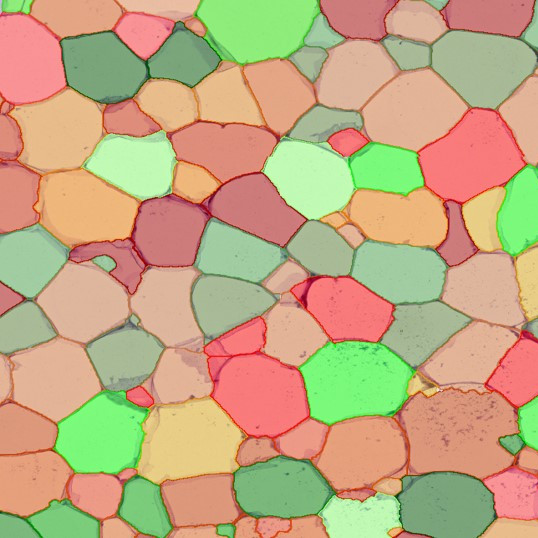
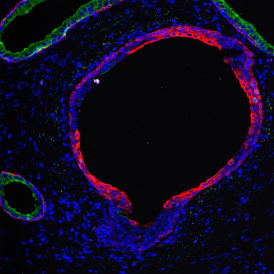
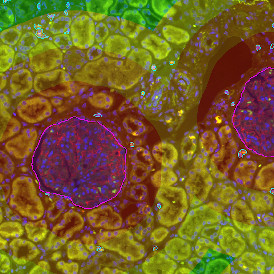
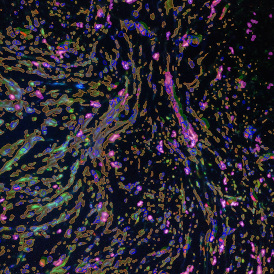
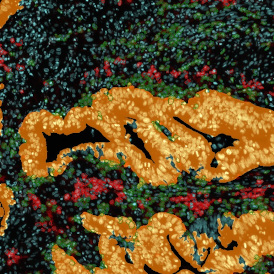
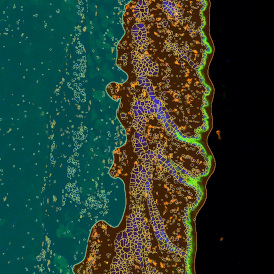
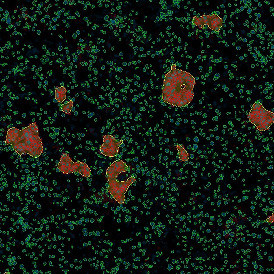
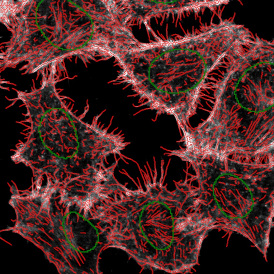
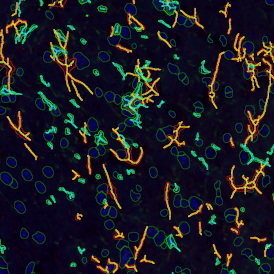
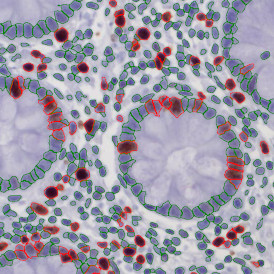
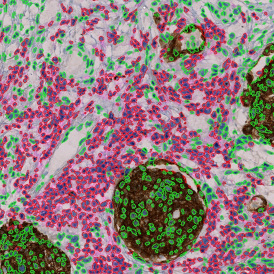
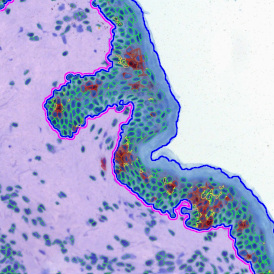
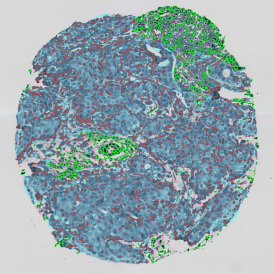
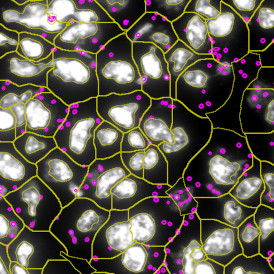
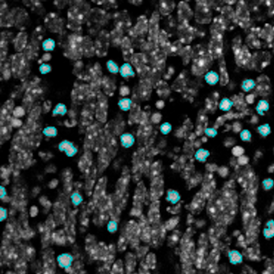
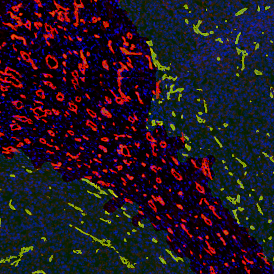
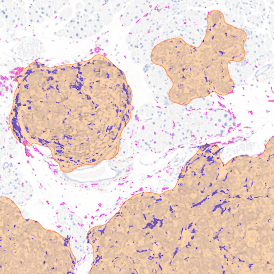
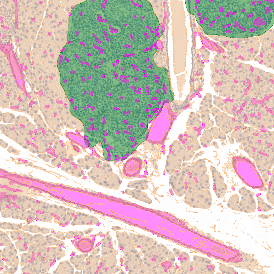
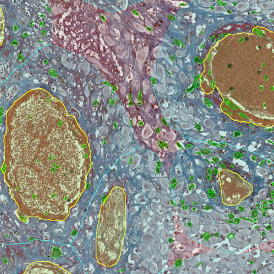
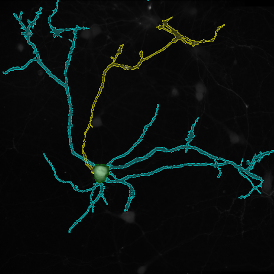
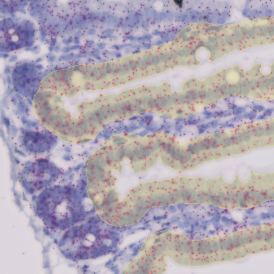
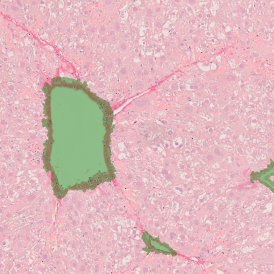
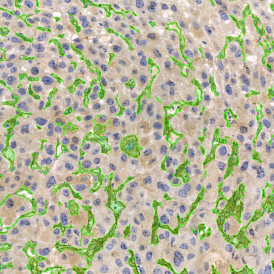
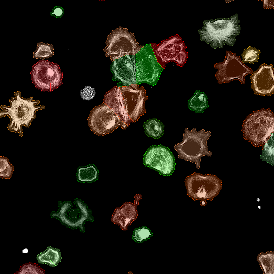
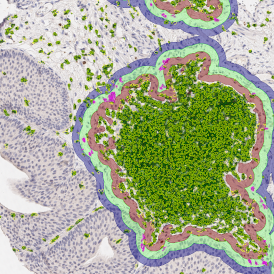
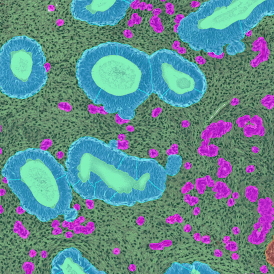
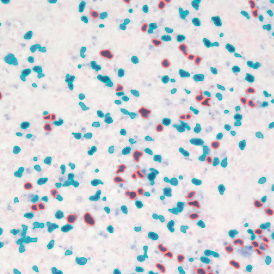
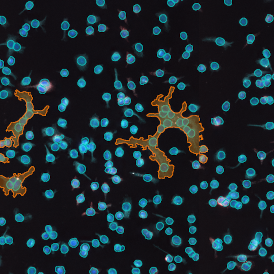
Immunofluorescence image analysis, as part of tissue cytometry, is an excellent method for understanding many immune changes and profiling tumor microenvironments.2 The workflow starts with sample preparation and staining of the target(s) of interest in cultured cells or tissue sections. Then, automated fluorescence whole-slide imaging can be performed and image analysis to detect nuclei or phenotypes can begin.
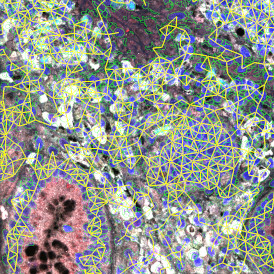
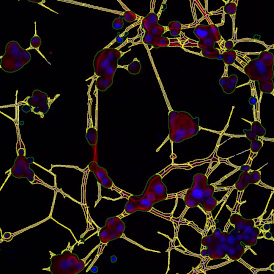
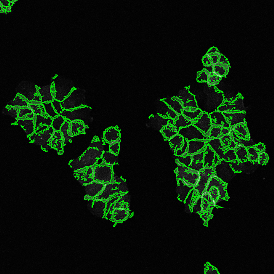
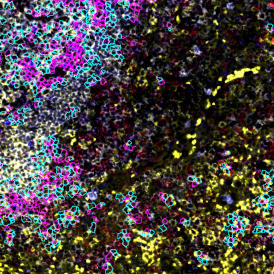
One of the great strengths of this method is that the technique can be multiplexed to examine multiple targets in the same tissue/experiment. This means the introduction of multiple markers onto a single tissue sample, typically emitting at different wavelengths so they can still be distinguished from each other, as well as multiple staining/imaging rounds (CycIF). While the use of multiplexing does introduce additional complexity into data analysis workflows, using automated imaging and analysis makes it possible to perform multiparametric analysis of a tissue section simultaneously.3
Some of the key challenges, particularly for multiplexed data sets, are dealing with large data sets, discovering automated ways to reconstruct images, finding computationally efficient ways to perform image segmentation, and trying to perform these immunofluorescence image analysis tasks in a standardized way. Other challenges concern the experimental protocols, optimization, spectral overlap, and finding the appropriate antibodies.
Optimization
There are several image-based cytometry techniques that are used in scientific research to study cells and tissue, including the two powerful methods of flow cytometry and tissue cytometry.
Optimization is key in immunofluorescence image analysis. Given the growing need within the life sciences for the detection of multiple markers, an increasing number of available protocols can help speed up the choice of antibodies and reduce the time taken to optimize the imaging procedure.4
Another consideration when performing experiments is whether the markers used undergo any photodamage or bleaching. It is important to choose the sample excitation conditions correctly in order to avoid this, as well as to minimize the creation of any autofluorescence in the tissue, which can appear as a large background.
Fluorophore markers should also be suitably bright to avoid creating issues with the signal-to-noise ratio. Excessive autofluorescence, common with plant samples, or scattering from the tissue itself can also cause issues with the signal-to-noise ratio as the desired fluorescence signal will be on a very intense background.
Other factors that should be considered when choosing the fluorophores and optimizing the immunofluorescence staining are spectral overlap and channel bleed-through. Having fluorophores with spectrally overlapping emission profiles can make it challenging to disentangle the different emission channels, even when filters isolate specific channels, as there can be unwanted emissions from another channel.
As well as choosing fluorophores with well-separated emissions and balancing the relative amounts of each fluorophore according to their brightness, another strategy is to use fluorophores that absorb at different wavelengths so that they are not excited simultaneously.
TF SPECTRA
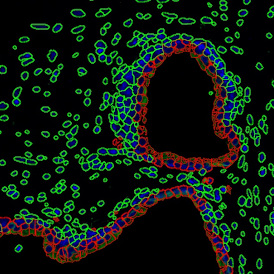
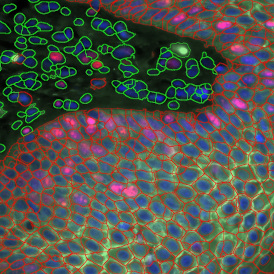
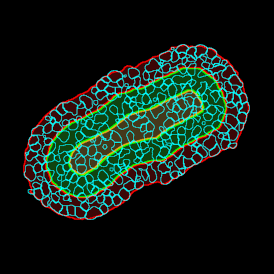
Despite the complexities of multiplexing for immunofluorescence image analysis, the ability to simultaneously record multiple channels has clear benefits in regards to the quality of information that can be obtained. The right tissue cytometer for image acquisition and data analysis can also make the process more straightforward.
TF SPECTRA is a multispectral imaging set-up capable of recording the emission of up to 8 markers when combined with our StrataQuest’s unmixing algorithms. Many possible artifacts, like channel bleed-through, can be potentially corrected with the right post-processing software.
TF SPECTRA is designed, among others, for high-end immunofluorescence image analysis and uses tunable liquid crystal filters to offer much greater flexibility in the wavelength filtering than is achievable with standard filters. Such filters can be tuned at speed to build lambda stacks to rapidly help with data correction.
Contact us today to find out how the high degree of automation on our TF SPECTRA could accelerate your immunofluorescence image analysis; and how combining this with StrataQuest for automated workflows could speed up the process of immunological studies, screenings, and therapeutics development.
References and Further Reading
- Wilson, C. M., Ospina, O. E., Townsend, M. K., Nguyen, J., Moran Segura, C., Schildkraut, J. M., Tworoger, S. S., Peres, L. C., & Fridley, B. L. (2021). Challenges and opportunities in the statistical analysis of multiplex immunofluorescence data. Cancers, 13(12). https://doi.org/10.3390/cancers13123031
- Lee, C. W., Ren, Y. J., Marella, M., Wang, M., Hartke, J., & Couto, S. S. (2020). Multiplex immunofluorescence staining and image analysis assay for diffuse large B cell lymphoma. Journal of Immunological Methods, 478(July 2019), 112714. https://doi.org/10.1016/j.jim.2019.112714
- Rojas, F., Hernandez, S., Lazcano, R., Laberiano-Fernandez, C., & Parra, E. R. (2022). Multiplex Immunofluorescence and the Digital Image Analysis Workflow for Evaluation of the Tumor Immune Environment in Translational Research. Frontiers in Oncology, 12, 1–15. https://doi.org/10.3389/fonc.2022.889886
- Thurin, M., Cesano, A., & Editors, F. M. M. (2020). Biomarkers for Immunotherapy of Cancer. Spinger Protocols. https://link.springer.com/protocol/10.1007/978-1-4939-9773-2_22