Spectral Imaging and Spatial Analysis of TMA Samples
The TissueFAXS tissue cytometers provide a streamlined and automated workflow for the scanning of tissue microarrays (TMAs), including preview scan, TMA detection, high acquisition scan, and visualization. In case samples are stained with several immunofluorescent markers, multispectral imaging combined with spectral unmixing technology might be useful to increase the number of markers visualized in the same sample. Here the liquid crystal tuneable filter is used in order to acquire the lambda stack, which is an image dataset using the same field acquired at different spectral intervals. This technology enables the removal of bleed-through from other channels. In addition, it is possible to identify autofluorescence and then subtract it from the other signals, making the final image clearer for further analysis.
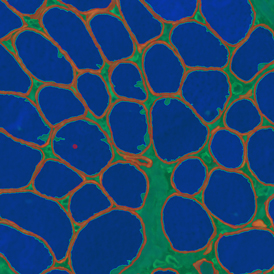
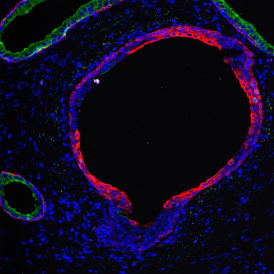
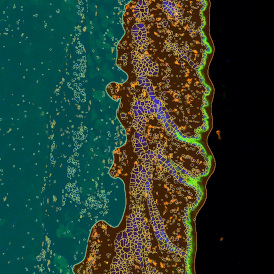
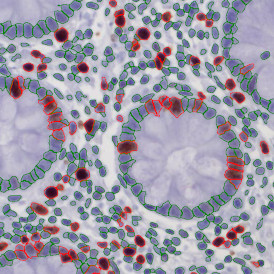
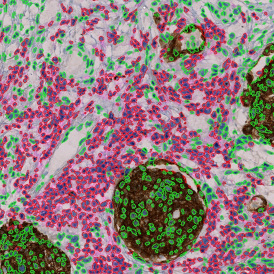
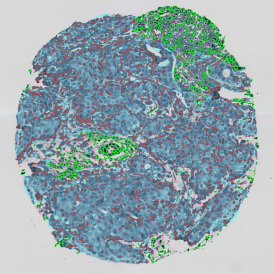
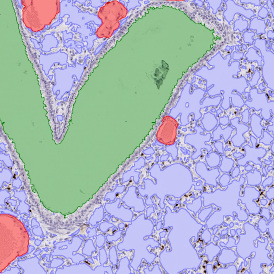
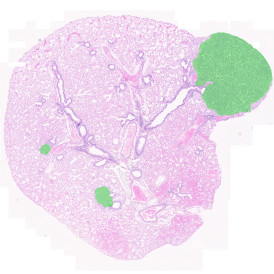
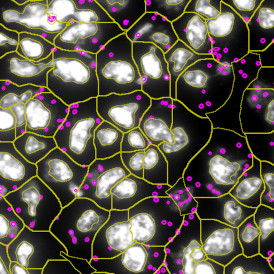
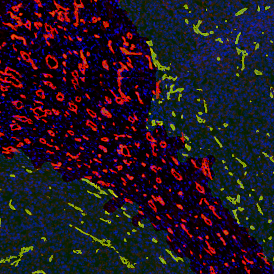
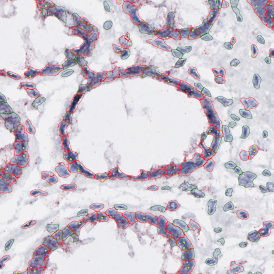
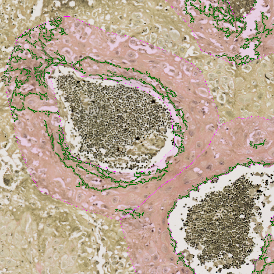
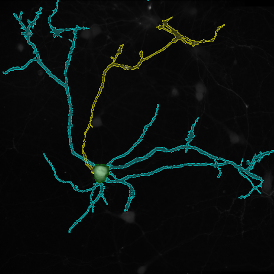
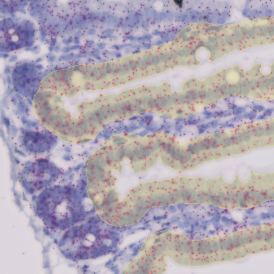
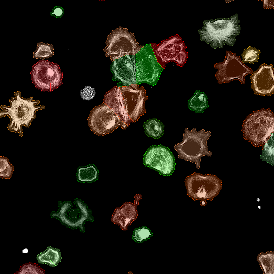
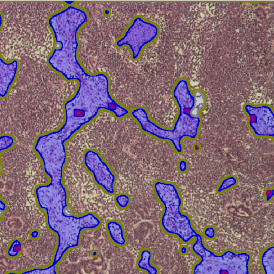
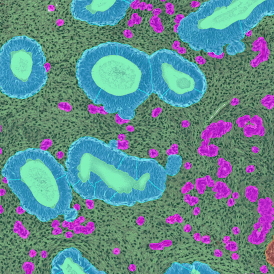
The example below shows a TMA core of inflamed colon tissue acquired with TissueFAXS Spectra, the multispectral system of TissueGnostics capable of multispectral imaging and unmixing. The image presents the visual difference between the original image on the left and the spectrally unmixed image on the right. The sample was stained for seven markers: DAPI (nuclei), CD4 (T helper cells), PD1 (participates in downregulating of the immune response), PD-L1 (ligand for PD1), CD68 (marker of monocyte lineage, macrophages), CD8 (cytotoxic T cells), and CK (epithelial cell marker).

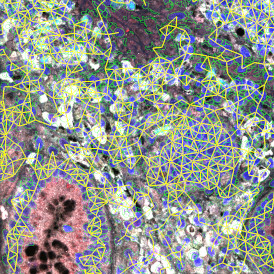
The aim was to decipher the immune microenvironment through the detection of single cells, in-depth phenotyping of the subpopulations, assessment of epithelium and stroma, and spatial analysis. The contextual image analysis software StrataQuest was used to analyze the sample.
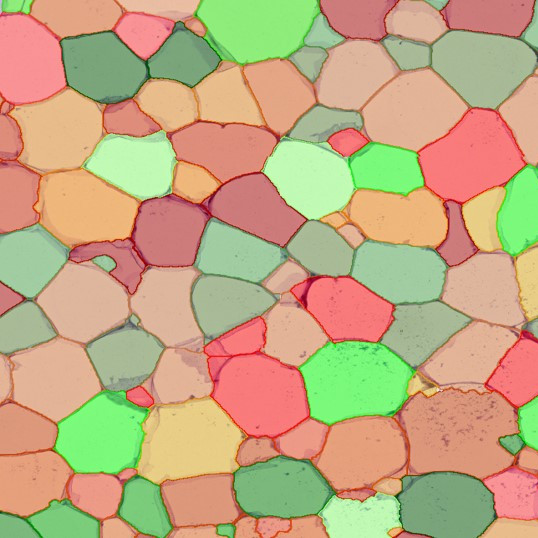
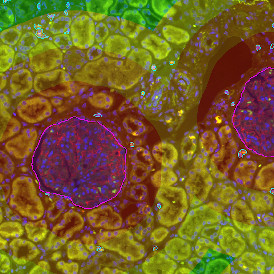
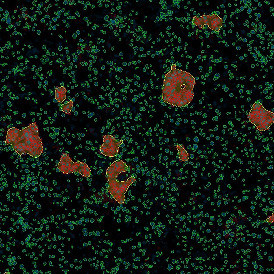
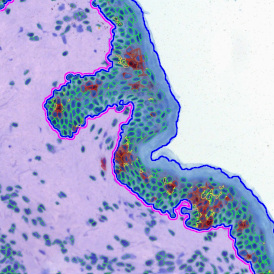
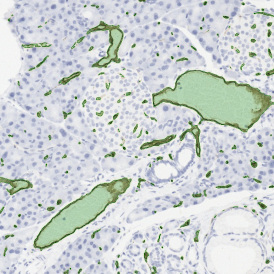
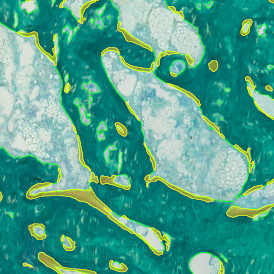
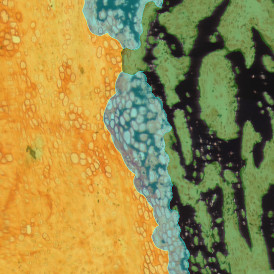
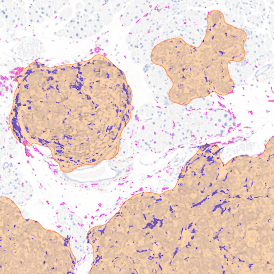
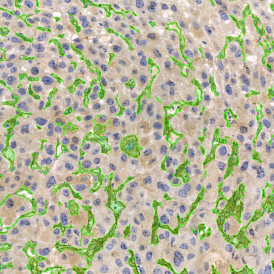
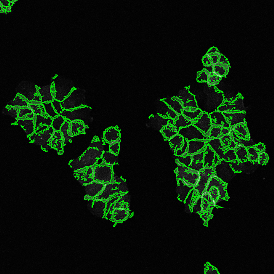
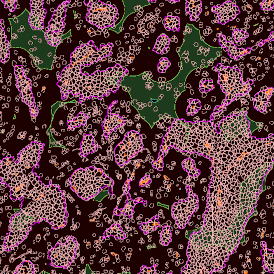
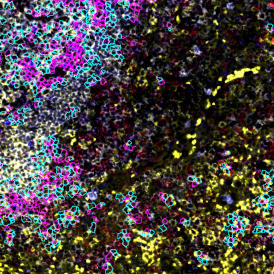
Image (a) is the unmixed image, and image (b) shows detected nuclei with a blue contour. Image (c) represents an example of the machine learning-based tissue classifier, available in StrataQuest, where the identified stroma is shown in green and epithelium in red overlay mask.

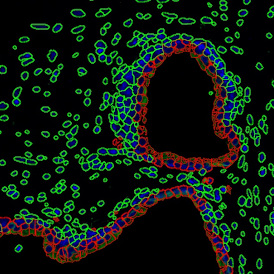
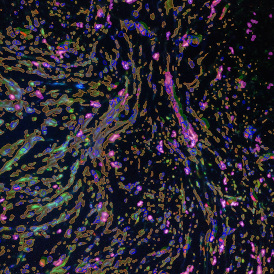
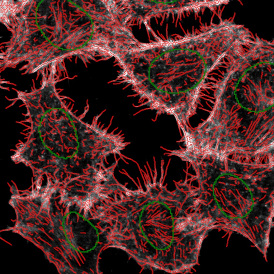
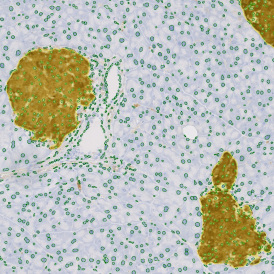
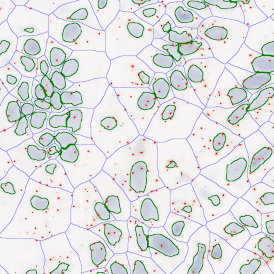
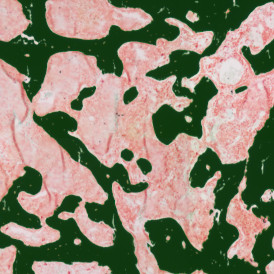
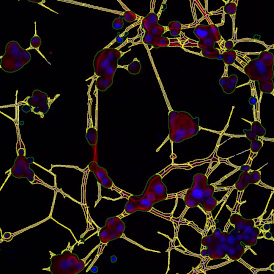
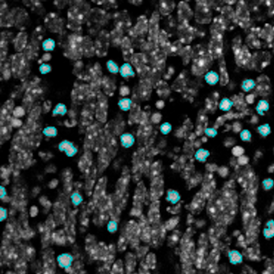
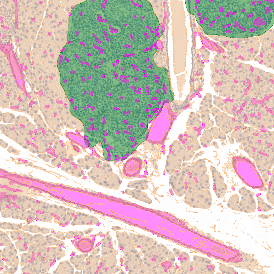
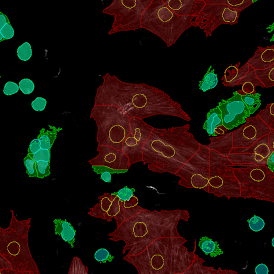
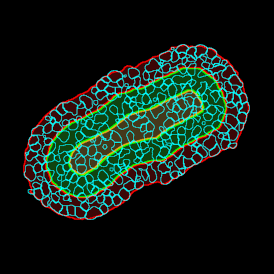
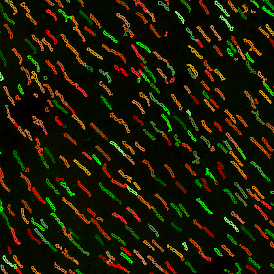
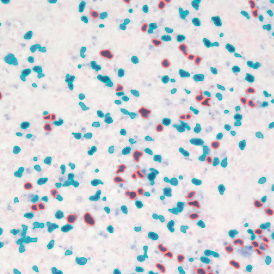
The next step was in-depth phenotyping of the individual immune cell subpopulations. The marker-stained cells were identified based on the intensity of the staining. StrataQuest also allows to show data for each marker separately. On image (d) CD4+ cells are visualized with a red contour, by using the backward connection. For validation, one can view the same image over a grey channel which shows signals from CD4+ cells only (e): marker-positive cells are shown in red, negative in green. Forward connection (f) can help to differentiate positive and negative cells: the same cell is marked on the image and corresponding scattergram, which delivers additional quantitative information. Like in Flow Cytometry, TissueGnostics image analysis software represents data in scattergrams and histograms.

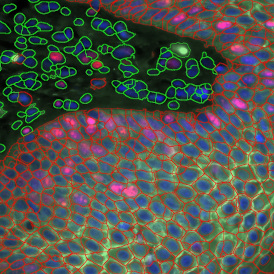
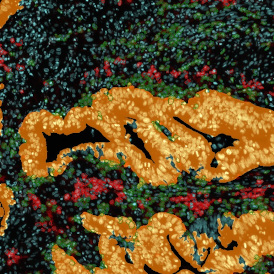
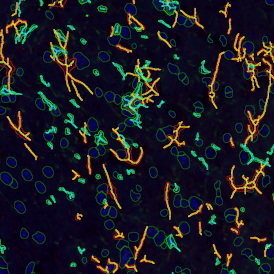
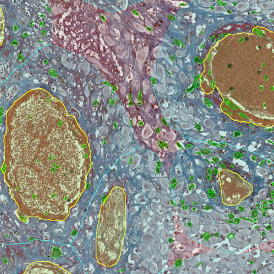
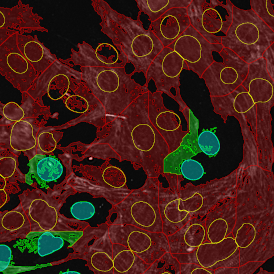
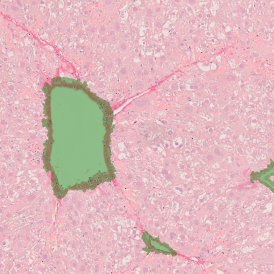
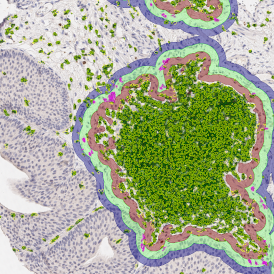
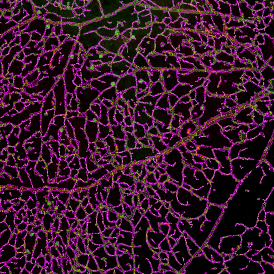
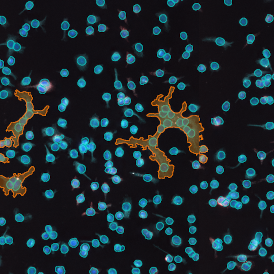
For many studies, the opportunity to obtain quantitative and qualitative data on the spatial scale is essential for the research. StrataQuest expands spatial analysis possibilities enabling cell-to-cell contact measurements (distances are definable): image (g) shows effector T cells (both CD4+ and CD8+) in orange contour, epithelial cells in light blue, and all other cell types in dark grey; while image (h) represents how the distance between immune and tumor cells are measured, in a range of around 30 µm.

In the end, all kinds of data (cell counts, intensities, proximities, areas, images, scatterplots, etc.) for each cell/TMA can be exported. All the obtained data can be further used for statistical analysis and data mining.
StrataQuest is TissueGnostics´ most evolved image processing solution for both brightfield and fluorescence images and is further capable of spectral unmixing, which is needed after multispectral imaging. If you are interested in performing spatial analysis or are looking for multispectral imaging solutions, please reach out to a member of the TissueGnostics team today.
Further sources: