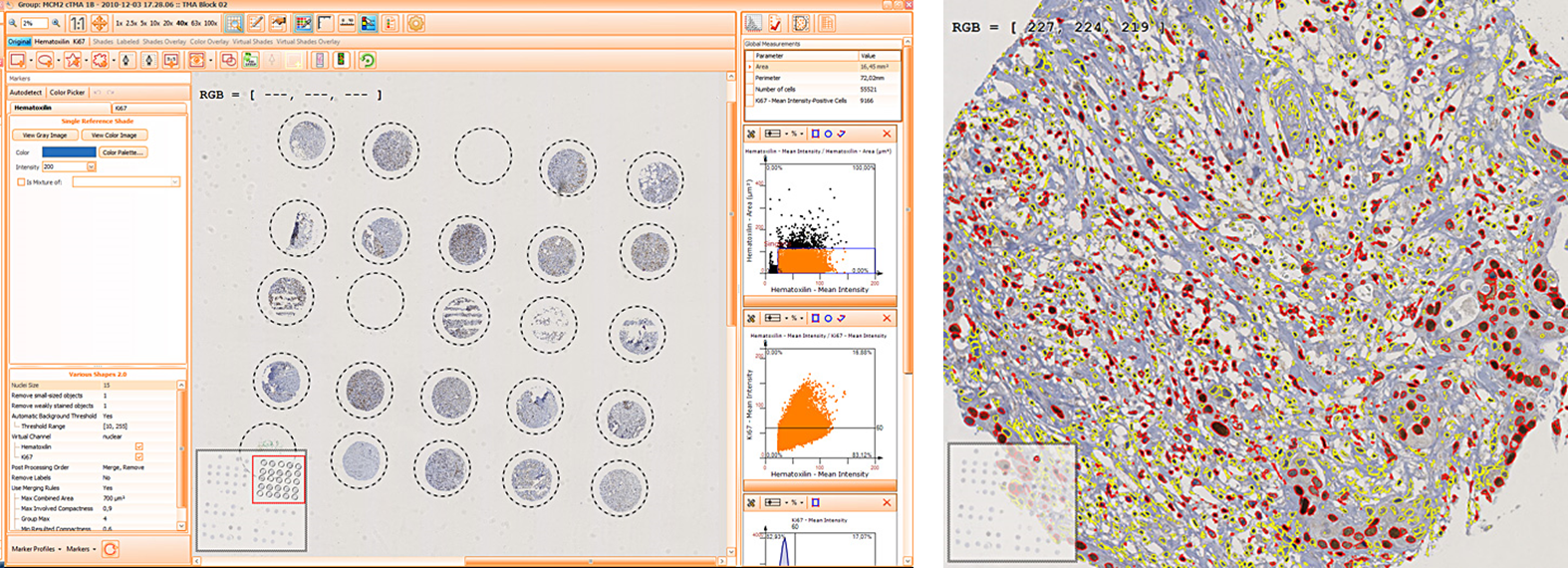
TISSUE CYTOMETRY TECHNOLOGY
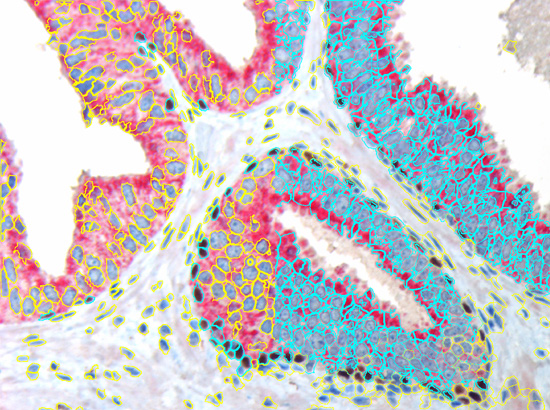
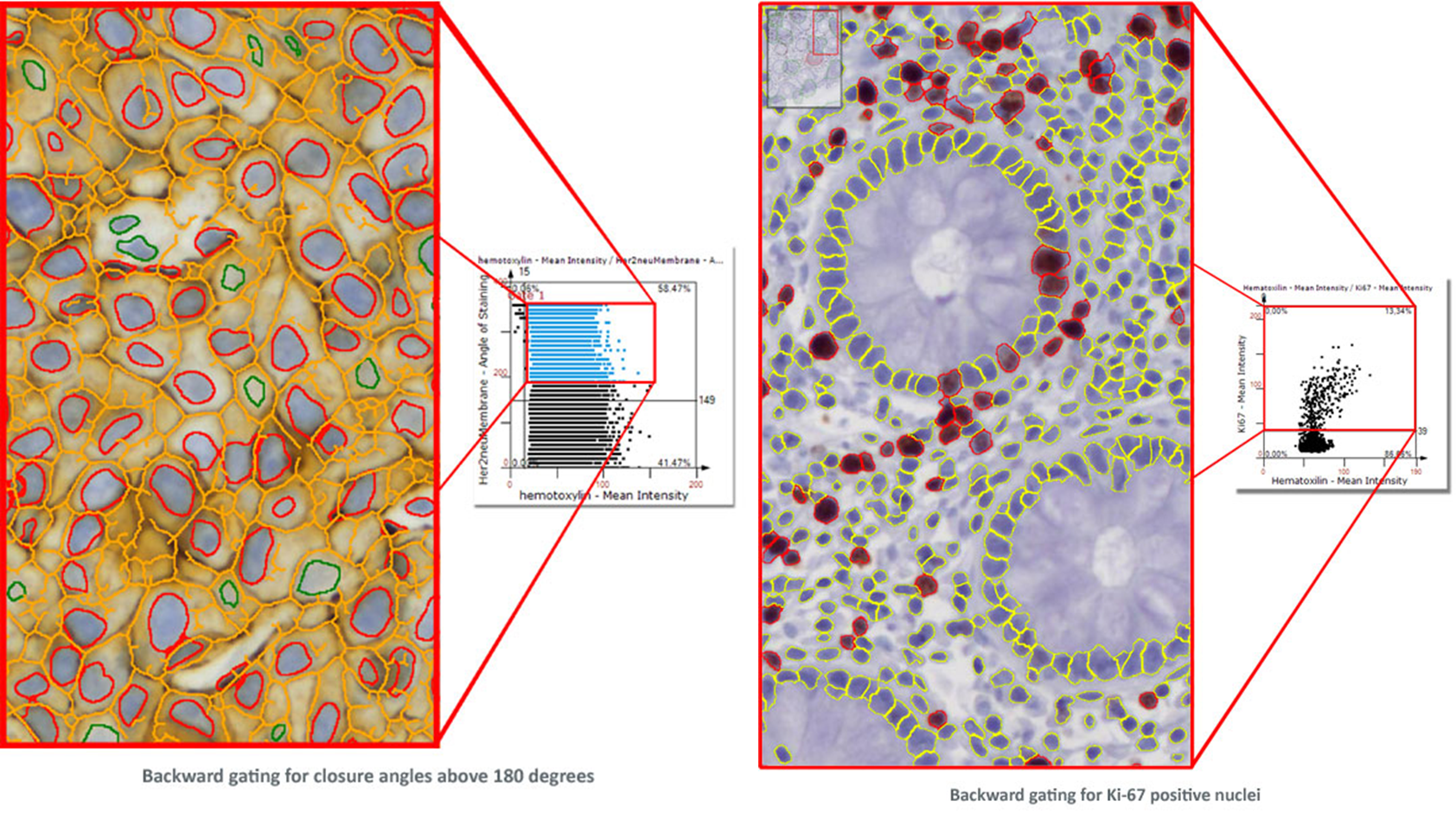
TissueGnostics introduced Tissue Cytometry technology into image analysis software more than a decade ago. While, at a first look, it may appear to be just another form of displaying data, there is much more to it. Combined with segmentation images it is also a simple and intuitive tool to visually validate results. TG calls this "Backward Gating". Backward gating can be done for any measured parameter on histograms and scattergrams. The following examples demonstrate the versatility of backward gating. In the left image backward gating is used to visualise cells in which the HER2/neu staining has a closure angle of more than 180 degrees. In the right image backward gating is used on segmented nuclei positive for Ki-67.
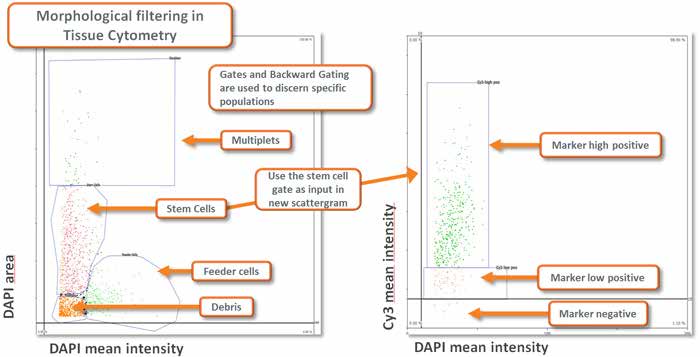
Morphological filtering using Tissue Cytometry technology combined with appropriate planning of the analysis provides a highly flexible and capable instrument for extracting specific data from a sample (see image below).


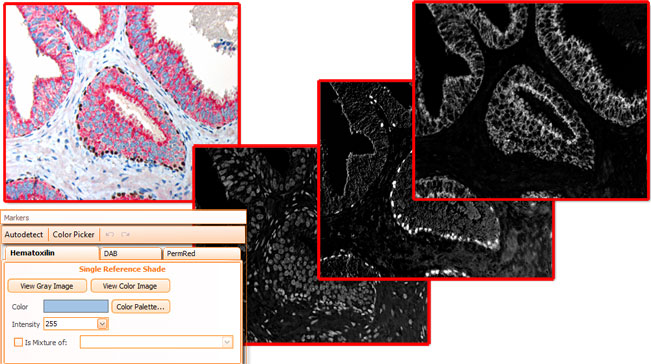 Brightfield images need to be separated into images according to their color components, HistoQuest provides two algorithms for color separation.
Brightfield images need to be separated into images according to their color components, HistoQuest provides two algorithms for color separation.