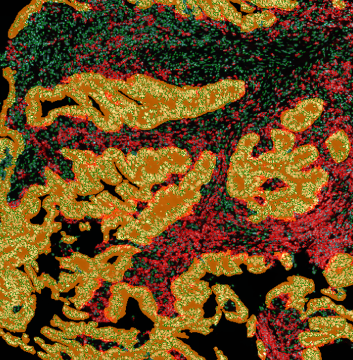
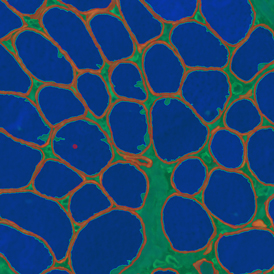
Adipocyte App: Measurement of Cellular Size of Adipocytes
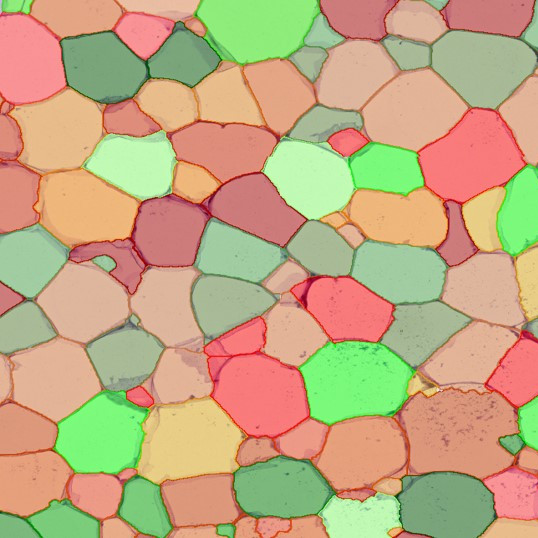
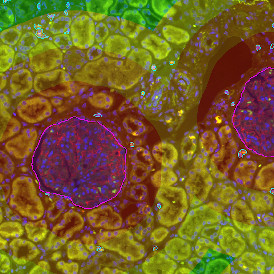
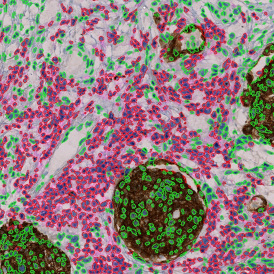
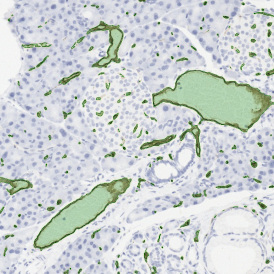
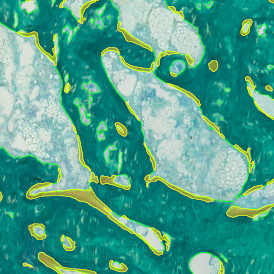
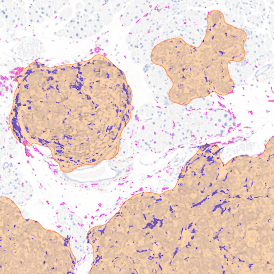
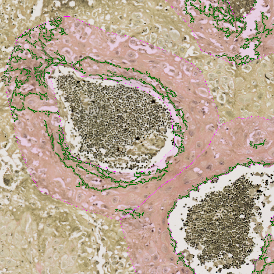
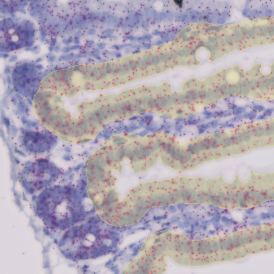
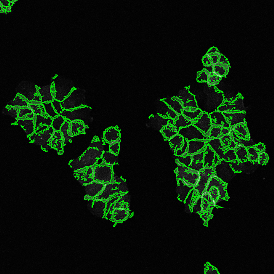
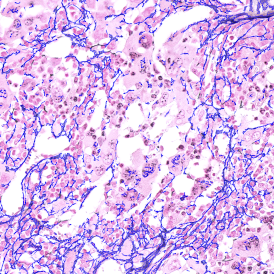
The StrataQuest-based Adipocyte App was applied to this sample with the aim to automatically detect (i) the number, (ii) the size, and (iii) the diameter of adipocytes within the given fat tissue. The challenge of analyzing the adipocytes is that the nuclei cannot be detected properly since they are either usually washed away during the H&E staining process, or are not visible. Therefore, the App detects the adipocytes using the membranes of the cells.
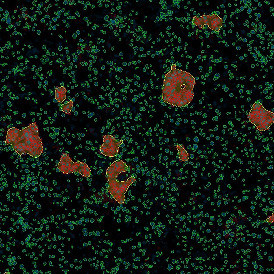
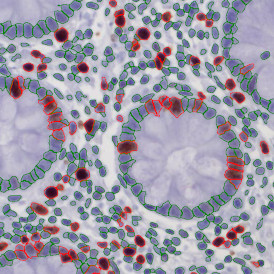
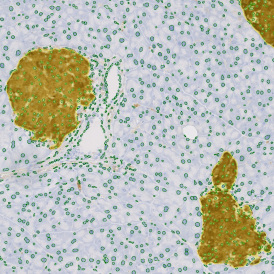
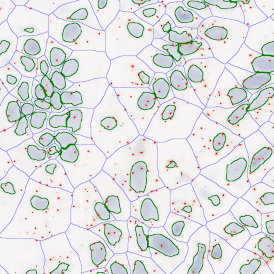
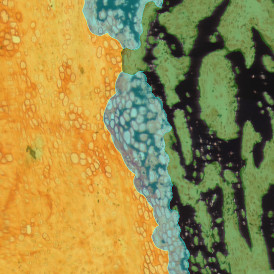
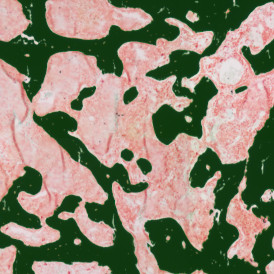
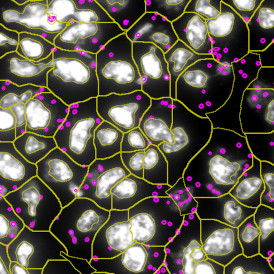
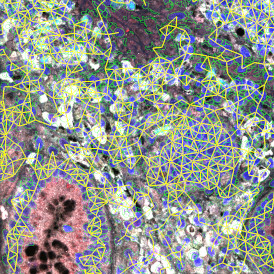
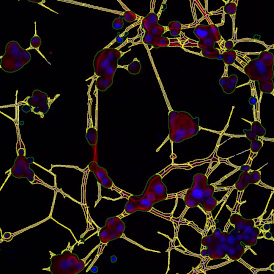
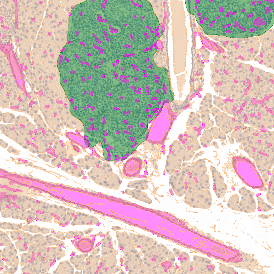
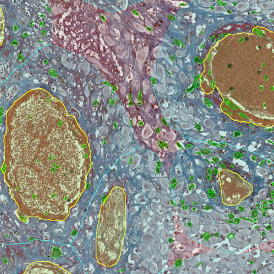
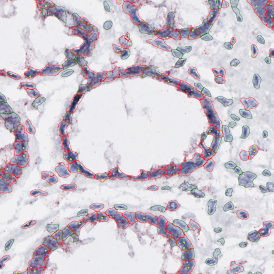
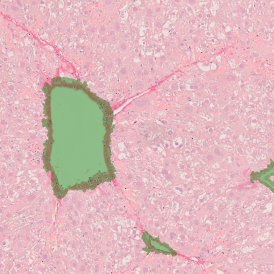
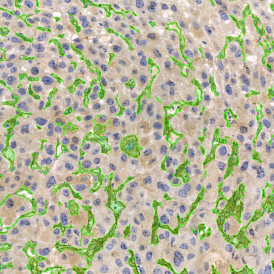
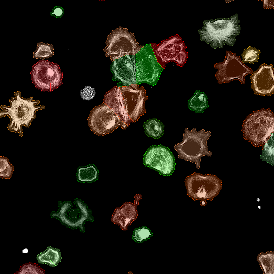
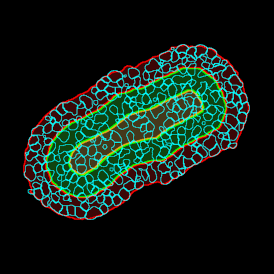
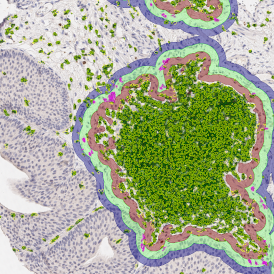
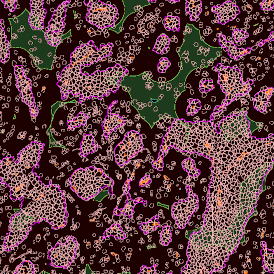
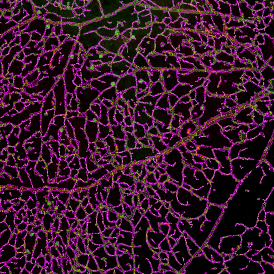
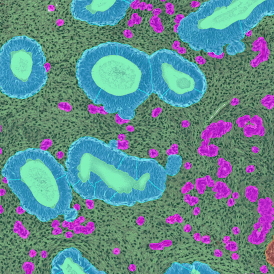
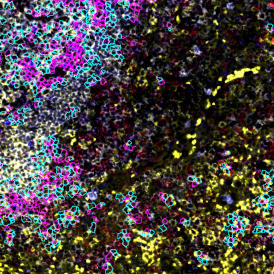
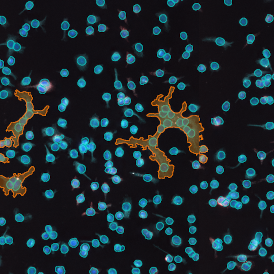
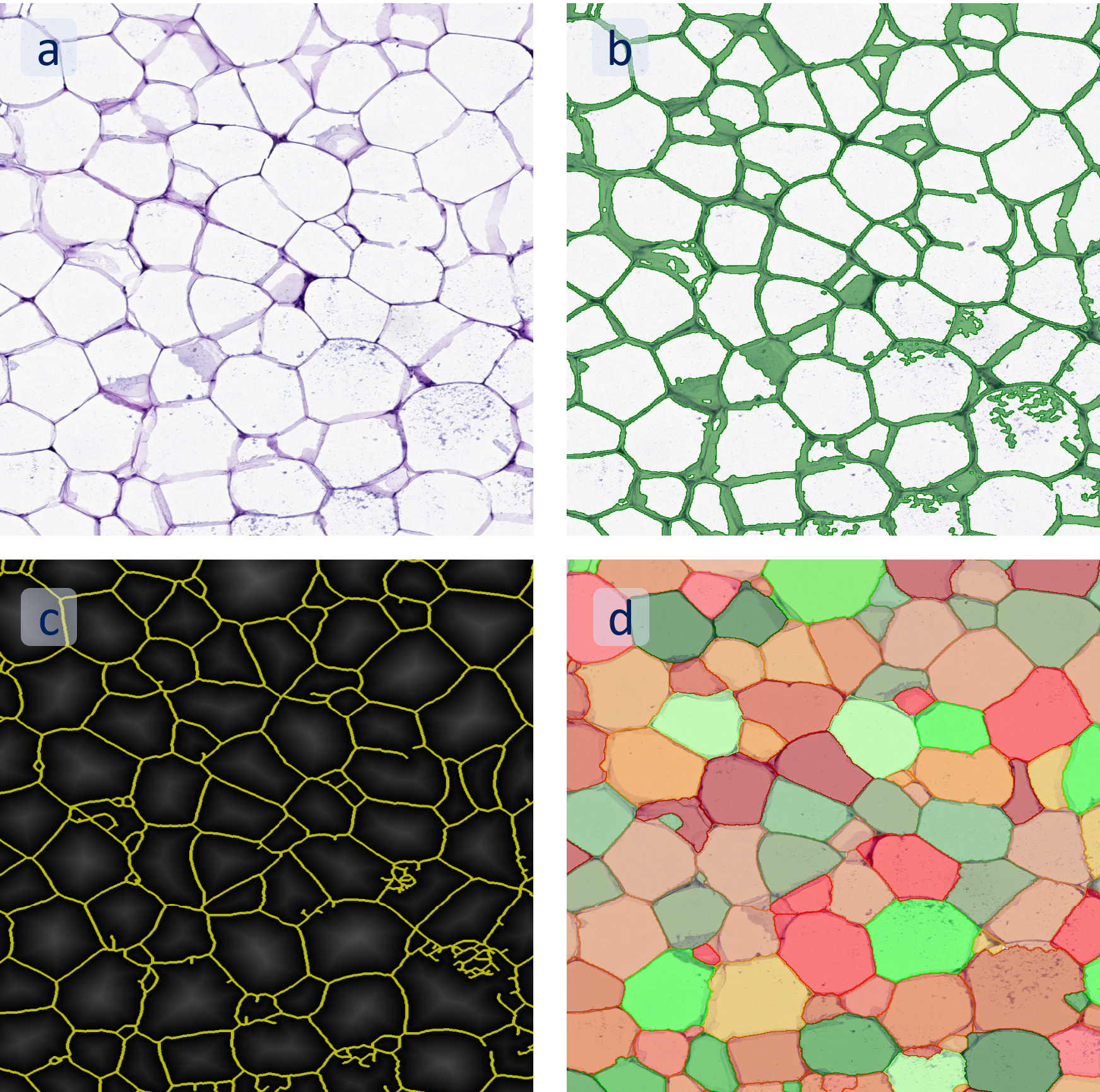
(a) Shows fat tissue stained with hematoxylin & eosin (H&E). The App first detects the membranes of the adipocytes and minor membrane tears are automatically closed (b). To detect the individual cells without nuclei, the App uses the detected membranes as a starting point and defines the cell by its distance from the membrane (c). Here the final result of cell detection is shown (d), cells are labeled in different colors for visualization purposes.
The App automatically eliminates adipocytes on the borders from the analysis which may influence the proper outcome of the analysis.
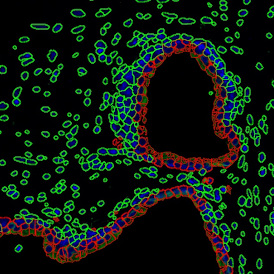
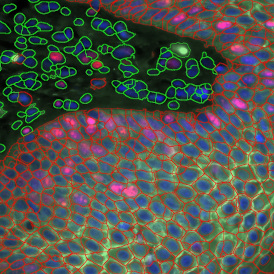
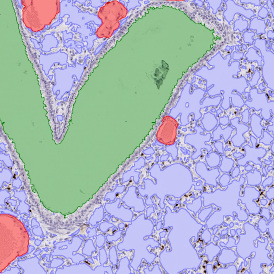
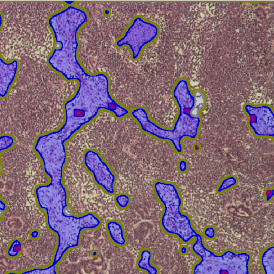
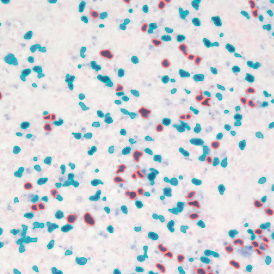
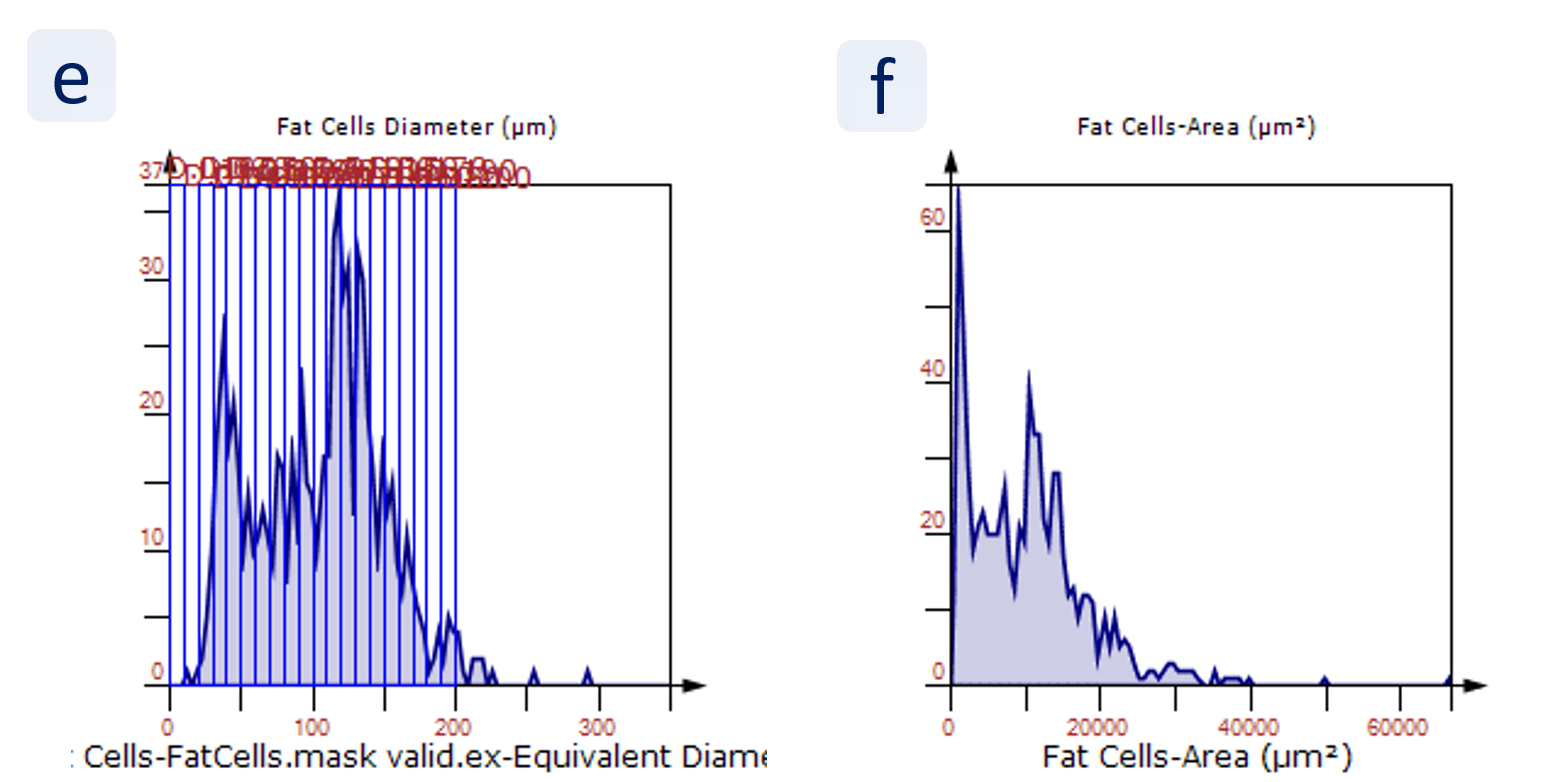
The results of the analysis are visualized in histograms and scattergrams. As shown in the scattergram below, the cells can be grouped according to their diameter (e) or (f) size.
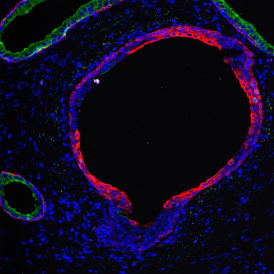
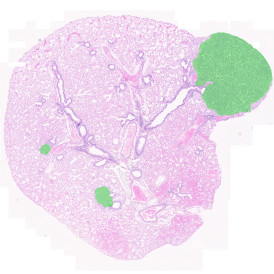
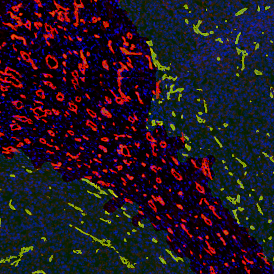
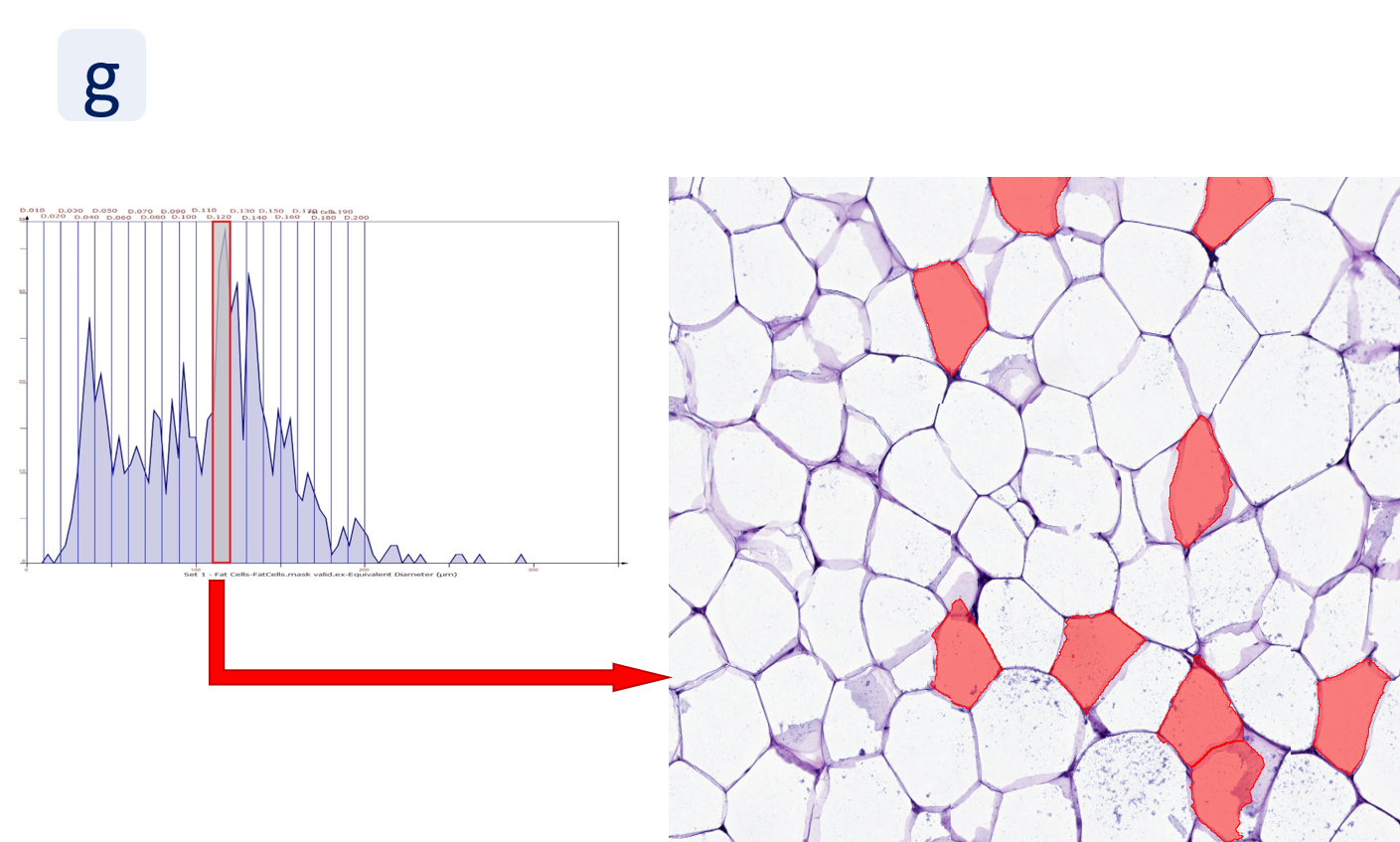
By selecting a gate of interest within the scattergram and using the backward connection feature, the cells from the selected gate are viewed directly in the sample in their native tissue environment (g). In this case, cells with diameters of 120µm to 129µm are shown in red.
The gained measurements can be exported from StrataQuest in CSV and /or Excel format.
StrataQuest Apps offer a streamlined and easy workflow for analyzing your samples. Look through our StrataQuest Apps and get inspired by the variety of biomedical image analysis applications. If you cannot find the perfect solution for your research question, please contact us. TissueGnostics offers the development of customized Apps for your specific analysis needs.