IF Dots App: Analysis of FISH
Fluorescence in situ hybridization (FISH) has been routinely used for decades in research and clinics to rapidly identify potential genetical changes within cells. This case study shows how TissueGnostics’ image analysis solution StrataQuest can be applied to evaluate FISH stainings. The most streamlined solution, the IF Dots App (runs within StrataQuest), can be applied to any analysis of dot-sized objects within the nuclei. Depending on the research needs, the following analysis can be combined with any other kind of analysis including AI-based tissue classification, phenotyping, spatial analysis, etc.
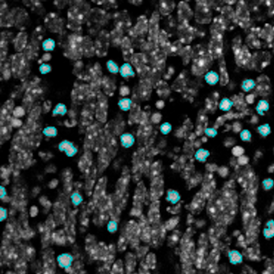
The aim of the following case study was to (i) identify individual nuclei and (ii) to assess how many FISH dots per individual nuclei are present.
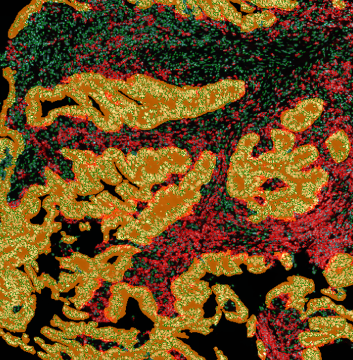
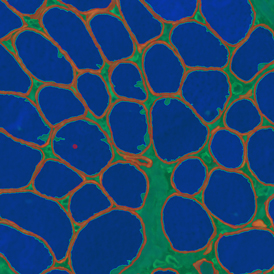
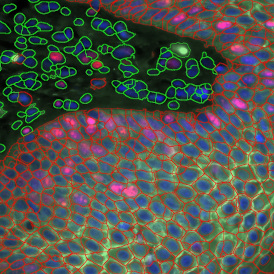
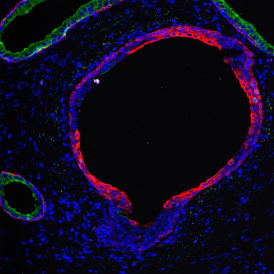
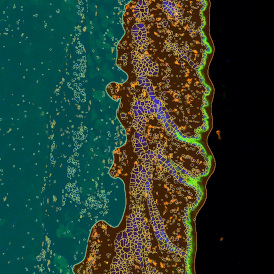
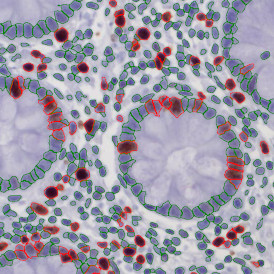
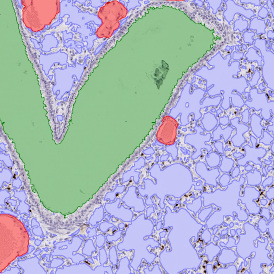
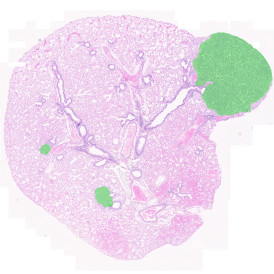
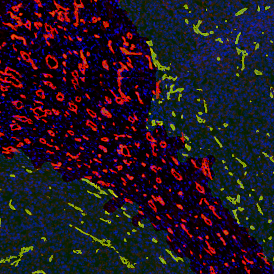
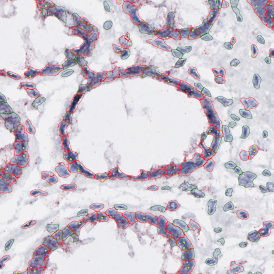
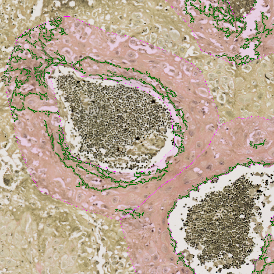
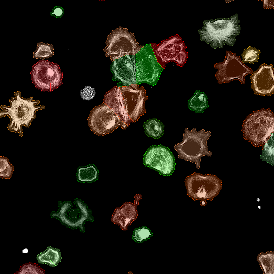
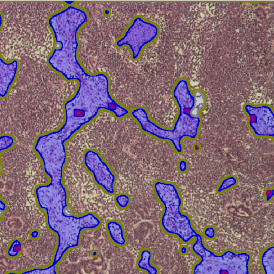
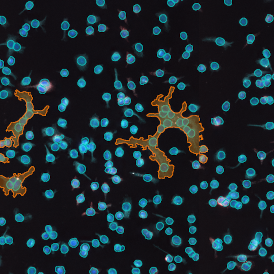
The image below shows an overview of a triple staining: the nucleus stained by DAPI (blue), and two FISH markers, the housekeeping gene (green), and HER2/neu (red).
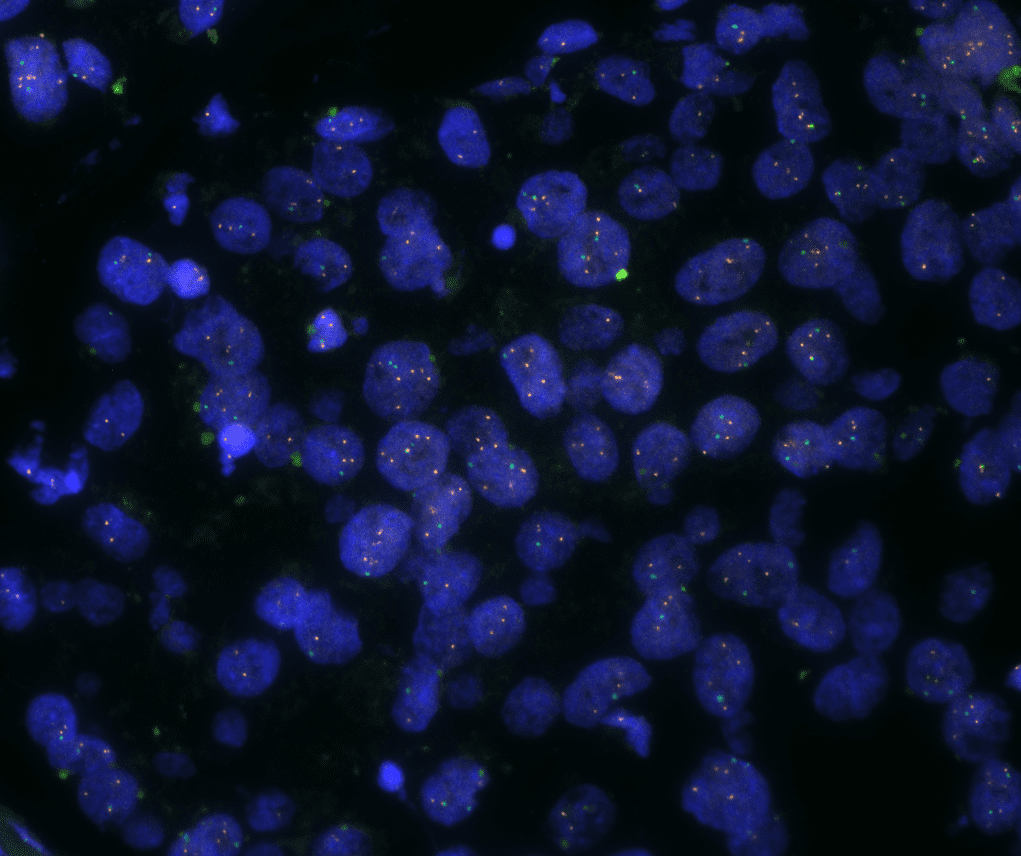
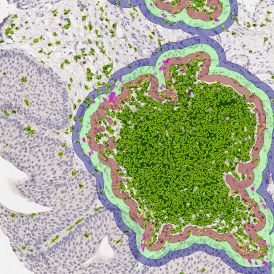
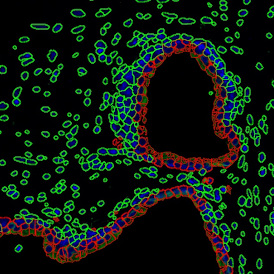
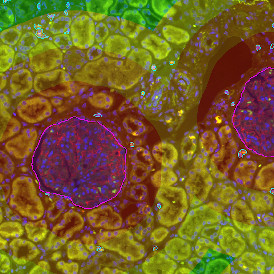
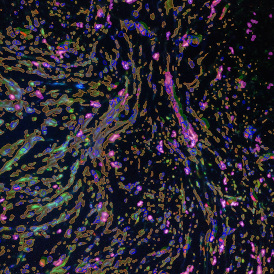
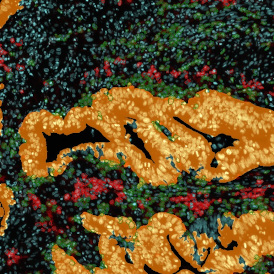
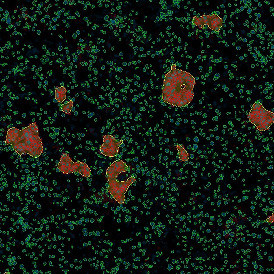
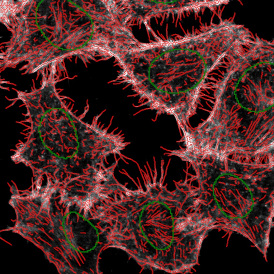
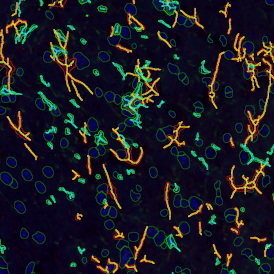
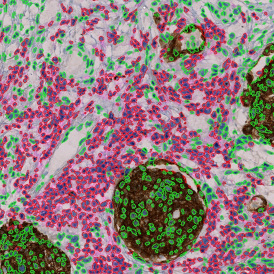
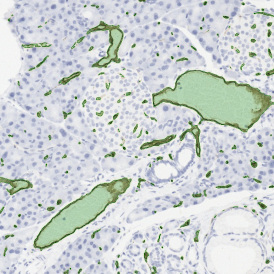
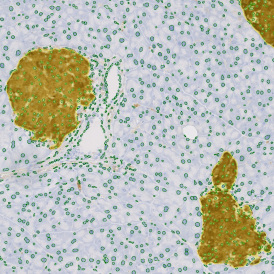
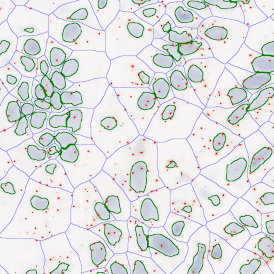
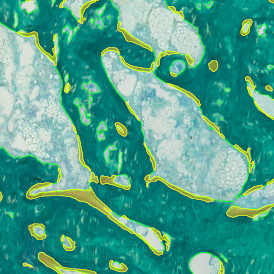
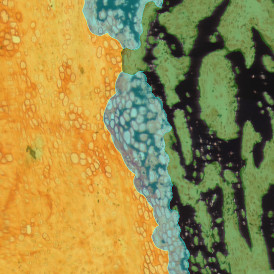
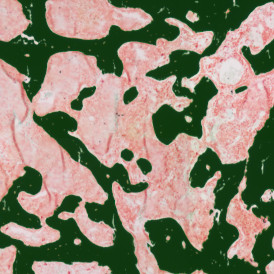
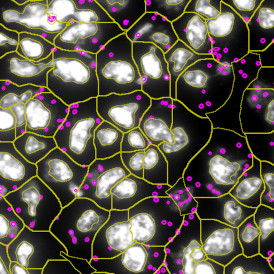
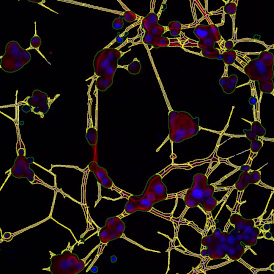
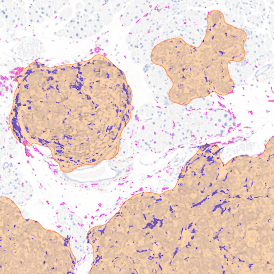
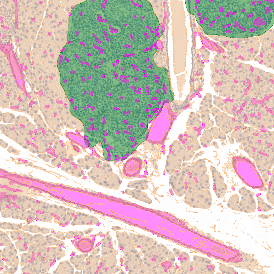
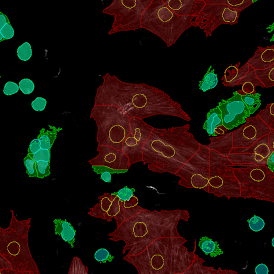
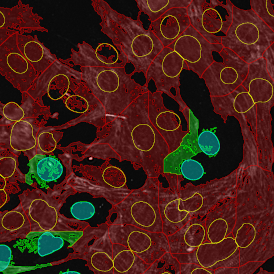
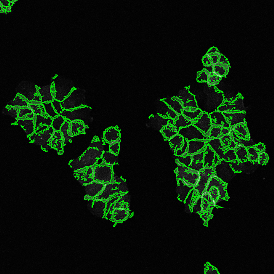
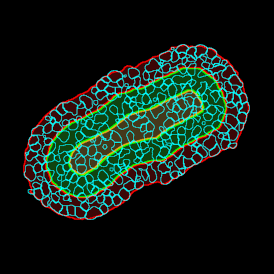
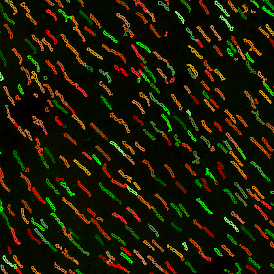
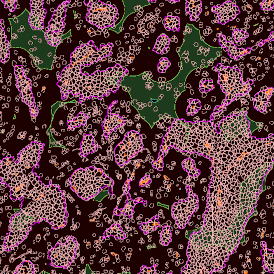
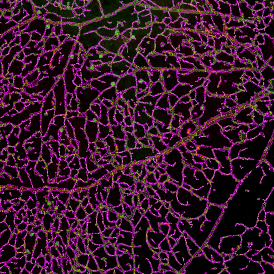
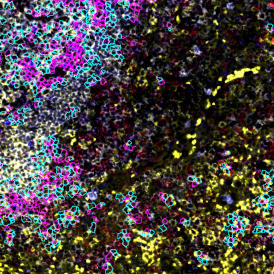
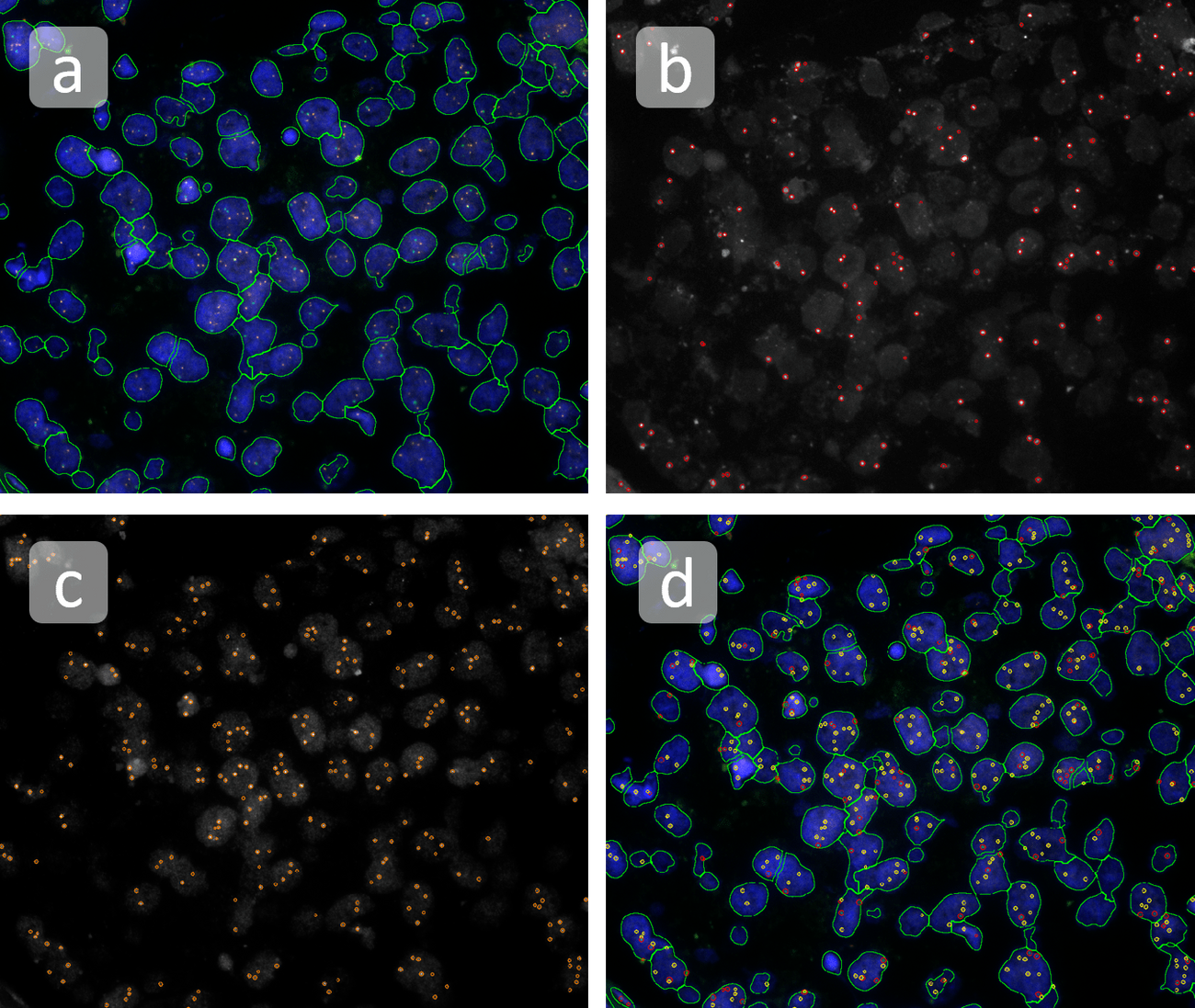
The automated analysis starts with nuclei segmentation (a), where nuclei are identified by the intensity of the DAPI staining (nuclei are marked by the green outline). In the next step, the FISH dots are detected: in red the housekeeping gene in FITC channel (b) and in yellow for the HER2/neu in Acridine channel (c). Then these masks can be combined, as seen in (d).
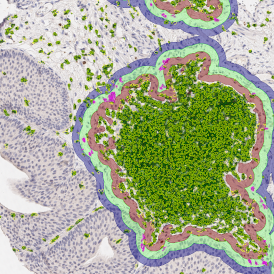
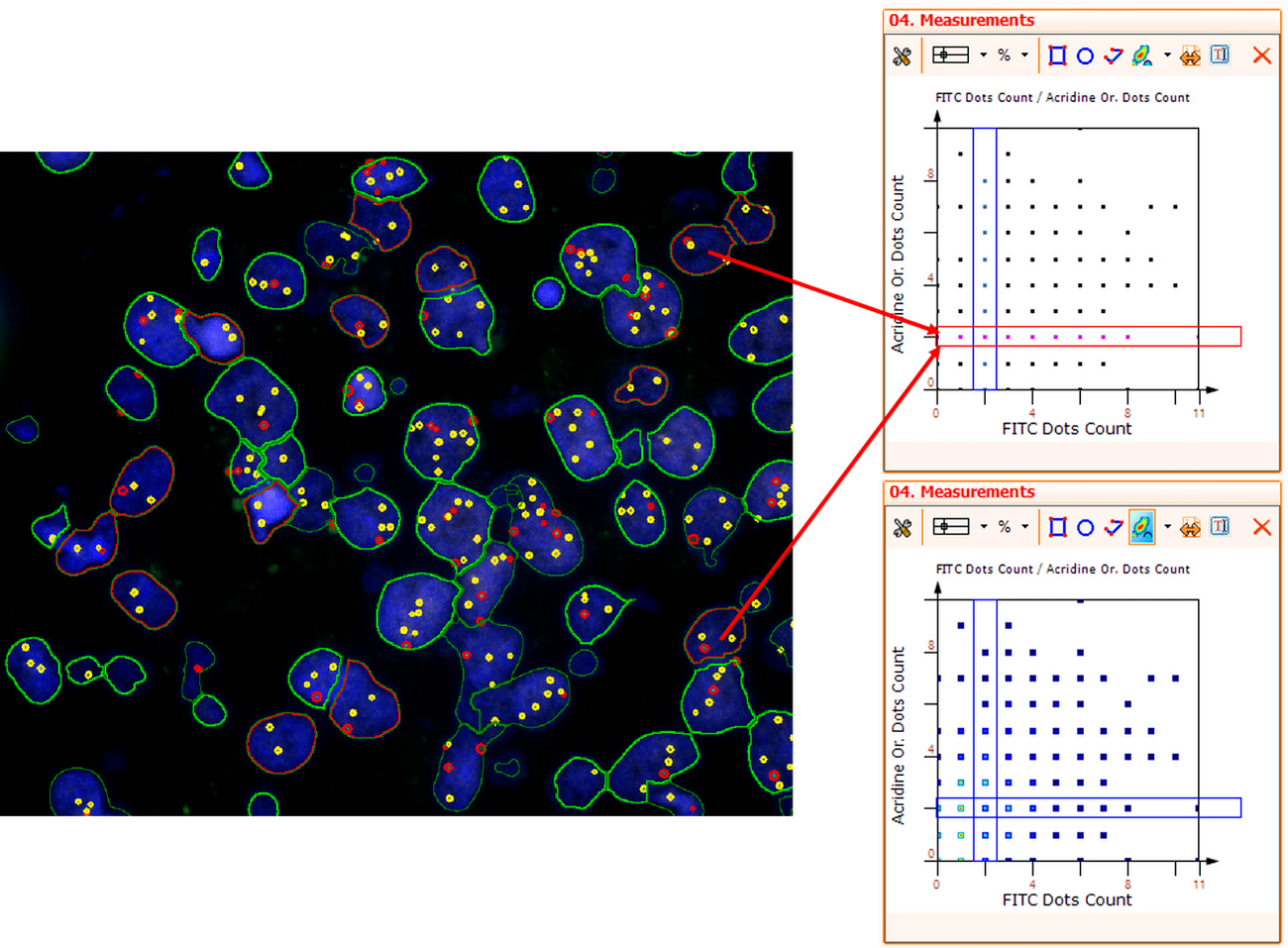
For further visualization of the analysis results, StrataQuest outputs the data in scattergrams. In the example below, the number of Acridine dots per nucleus (y-axis) is plotted against the FITC dots per nucleus (x-axis). The identified nuclei were gated for the condition “2 acridine dots per nuclei”, where the number of FITC dots can vary, and selected cells are shown in red contour via backward connection. Scattergrams showing the number of detected nuclei with each dot number combination are useful, especially with the Heat Map function enabled (bottom) which indicates those groups with a higher quantity of nuclei. To gain quantitative data for statistical analysis, the results can be immediately exported to Excel, CSV, and PDF formats.
FISH analysis is only one example of StrataQuest´s capabilities. If you are interested in how StrataQuest streamlined image analysis solution can enhance your research today, please reach out to a TissueGnostics team member for a demonstration.
Get to know more about FISH applications combined with TissueGnostics solutions in our blog posts:



 TissueFAXS Q systems are also available in scan only configurations.
TissueFAXS Q systems are also available in scan only configurations.