IF Dots APP: FISH
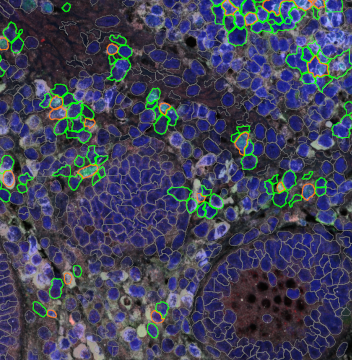
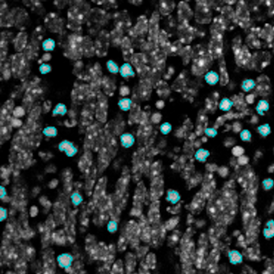
This example shows how the IF Dots APP can be applied to FISH evaluations. The APP can also be applied to any analysis of dot sized objects, in conjunction with all other StrataQuest features. For FISH evaluation the recommended workflow is to run the dot algorithm on all dot stainings and the to select suitable nuclei by their large size and high compactness.
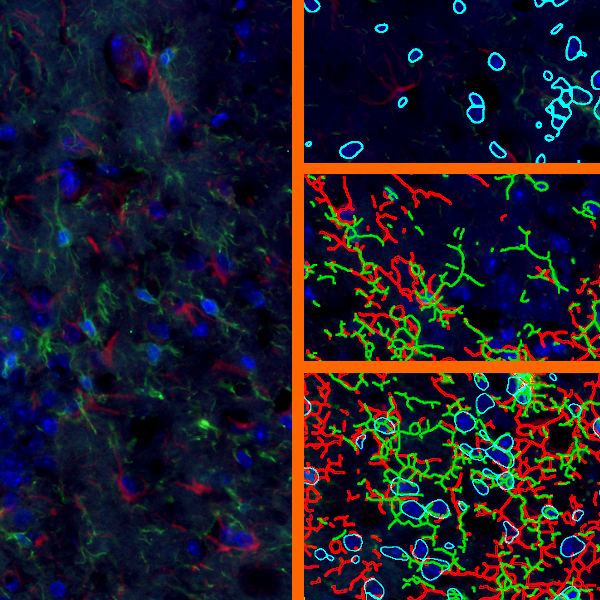
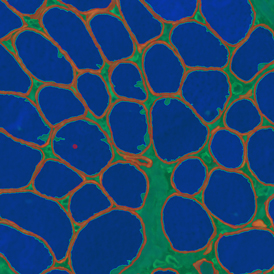
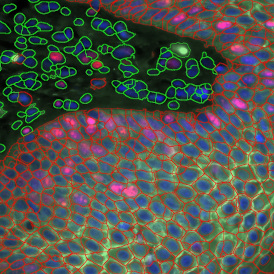
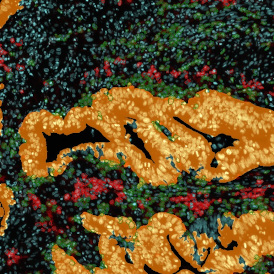
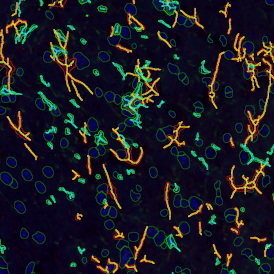
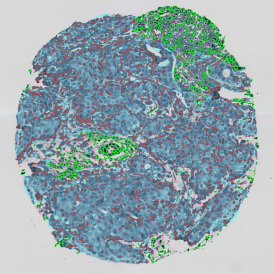
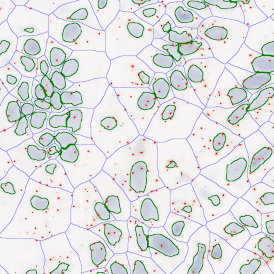
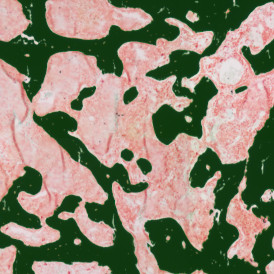
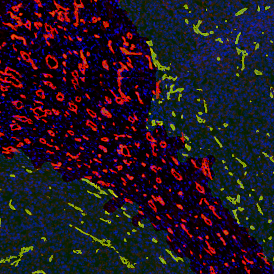
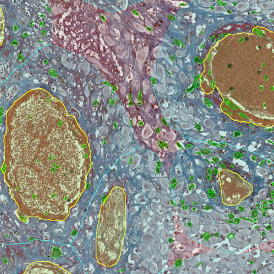
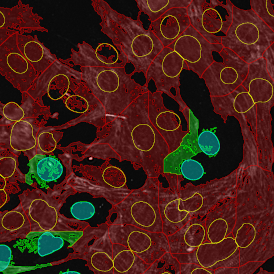
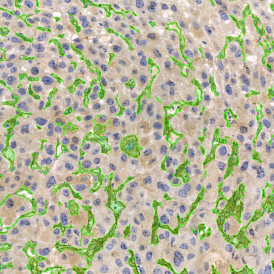
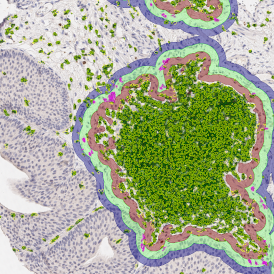
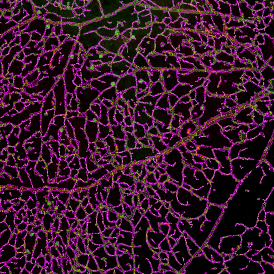
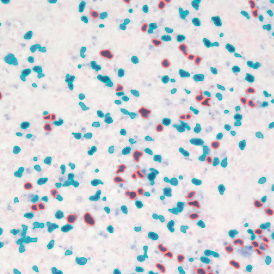
Overview of triple staining showing the nucleus in DAPI (blue), the housekeeping gene (green) and HER2/neu (red).


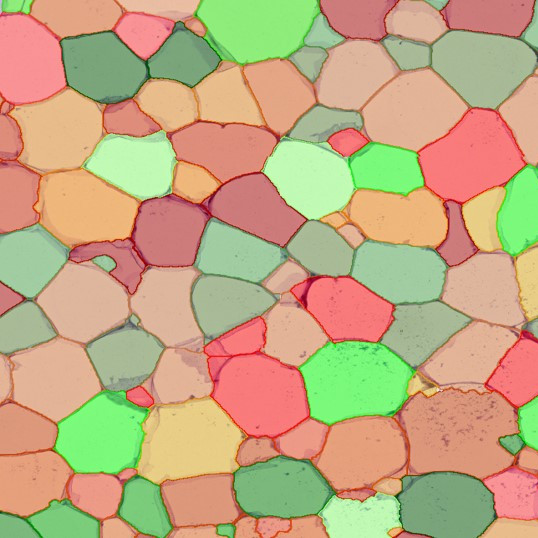
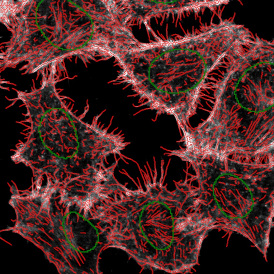
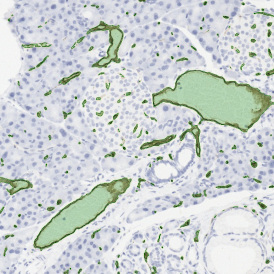
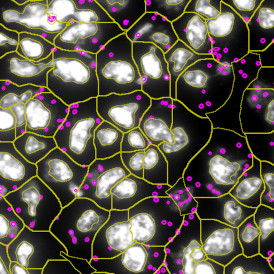
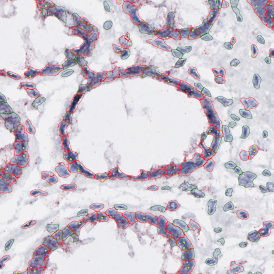
Selection of suitable nuclei by size and compactness.


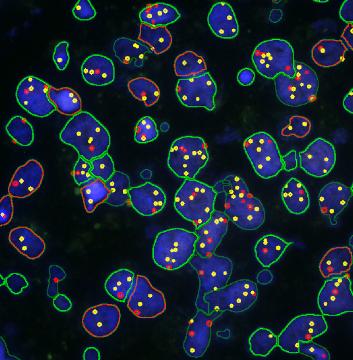
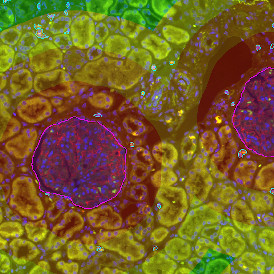
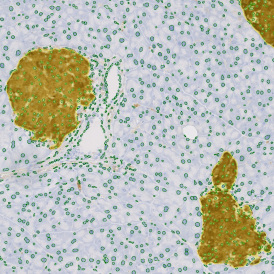
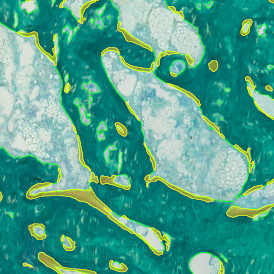
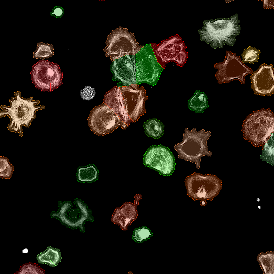
Detection of green housekeeping gene signal is shown in yellow in the right image and the detection of red HER2/neu signal in yellow in the left image.


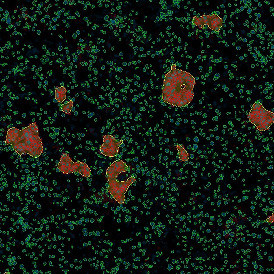
Cells with a 3 to 2 dots ratio (only one in this image) are shown by backward gating from the "3 x 2" gate.

FISH dot can be displayed and exported in two ways, as shown below. Scattergrams showing the number of cells with each dot number combination is useful, especially with the Heat Map function enabled which shows those groups with more cells. For the immediate export of data for pathology applications a special table view showing 16 parameters is provided (shown partially on the right side) which offers export to Excel, CSV and PDF formats.



IHC Immune Status in situ APP: Tumor immunology
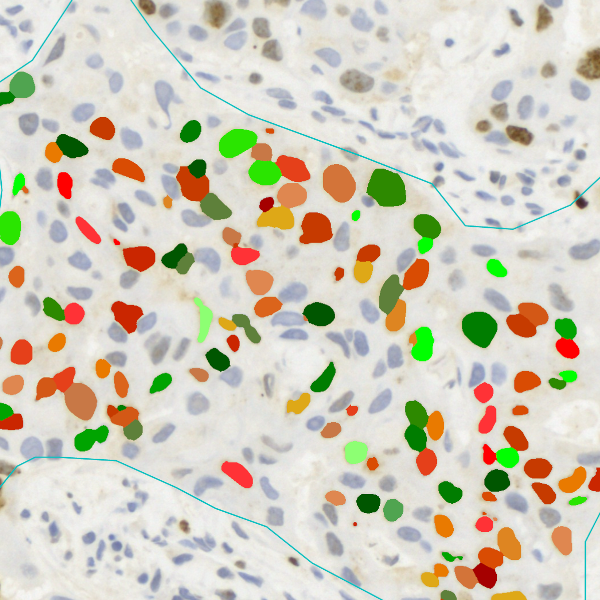
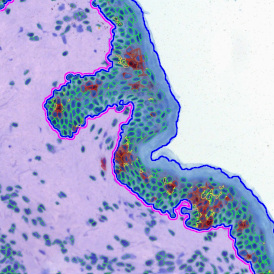
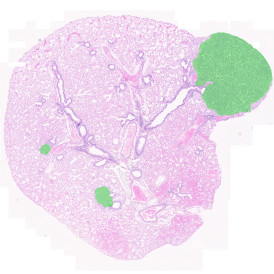
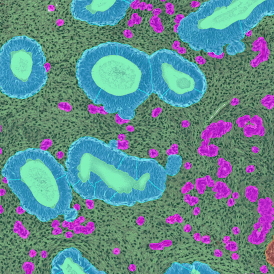
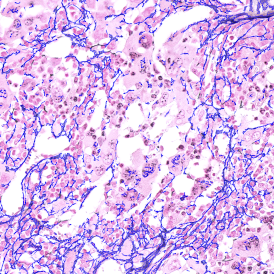
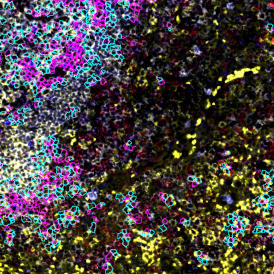
The IHC Immune Status in situ APP is ideally suited for immunological investigations due to its capability to measure the distance of single cells from tissue metastructures. In this example this is applied to bladder cancer Tissue Microarrays (TMA). In this TMA core DAB stained CD3+ lymphocytes (brown) are evaluted in the context of their distance from automatically detected tumor regions as well as from large and small lymphocyte clusters.
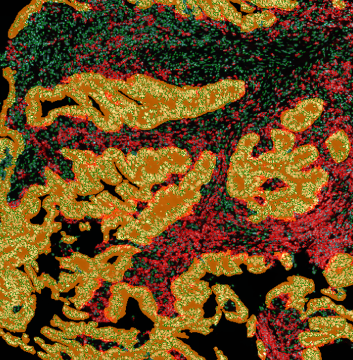
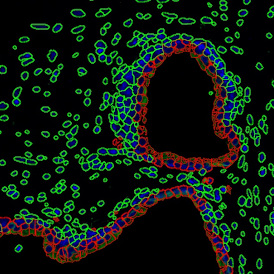
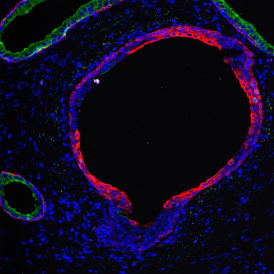
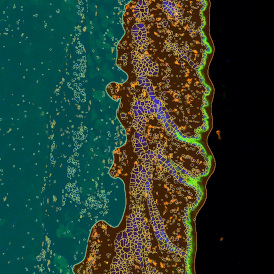
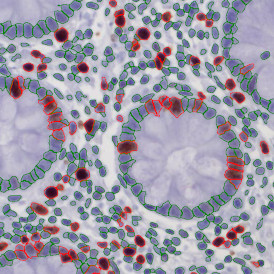
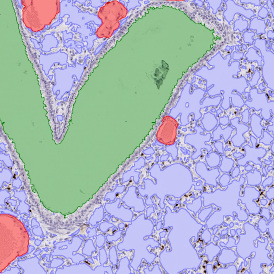
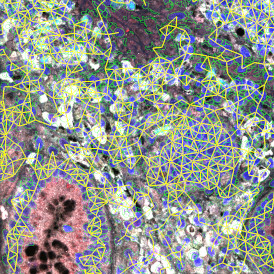
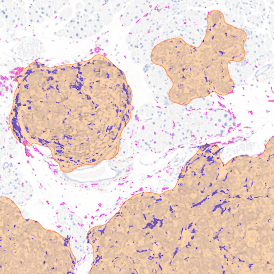
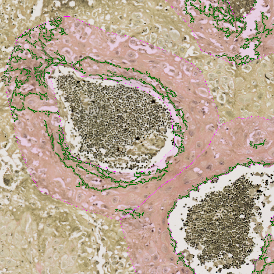
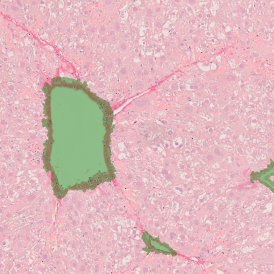
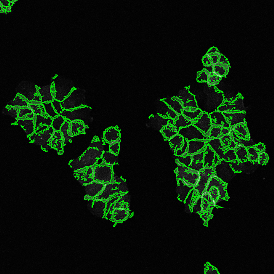
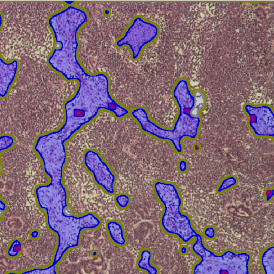
TMA core with tumor detection and tumor proximity ranges


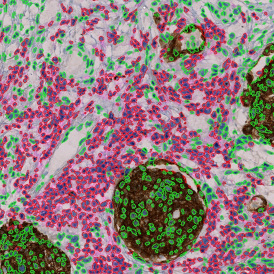
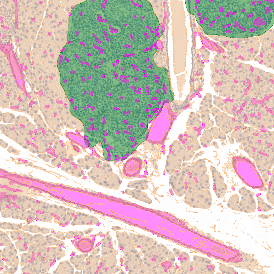
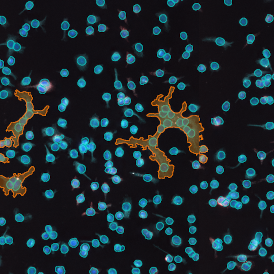
TMA core with lymphocyte cluster detection and lymphocyte cluster proximity ranges


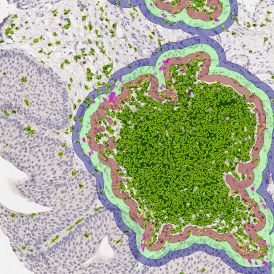
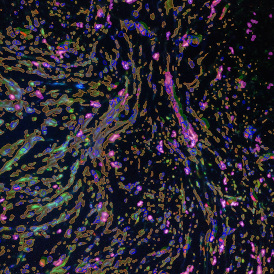
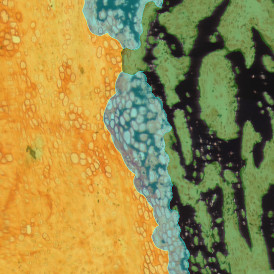
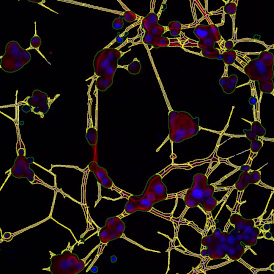
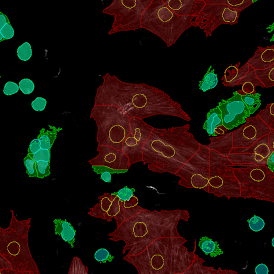
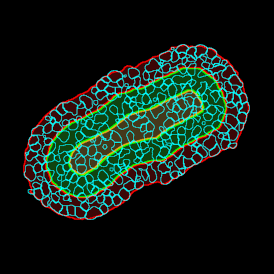
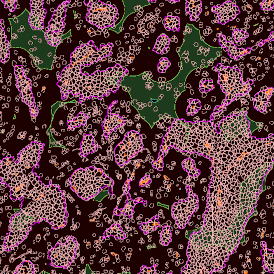
Lymphocytes (CD3+, purple contour) within tumor proximity


Lymphocytes (CD3+, purple contour) within lymphocyte cluster proximity


Sample courtesy of Dr. Kathleen Williamson, Centre for Cancer Research and Cell Biology, Queen’s University, Belfast