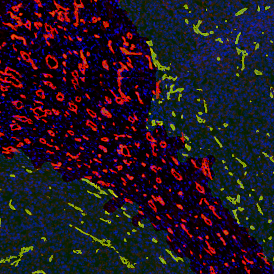
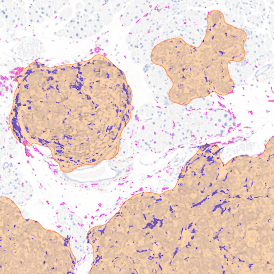
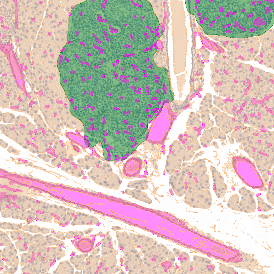
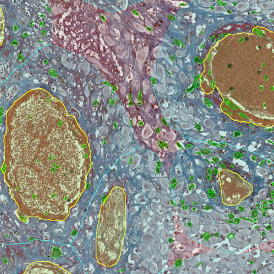
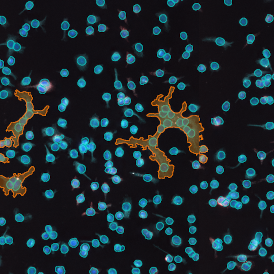
IHC META.CELLS
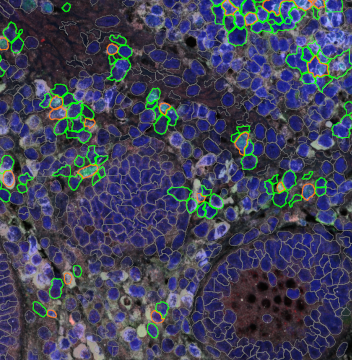
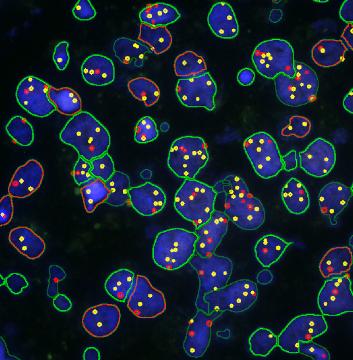
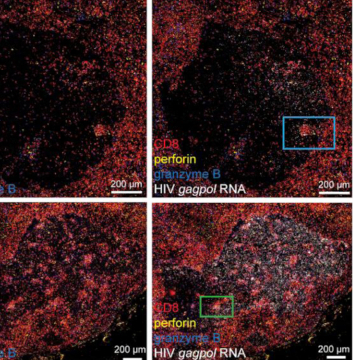
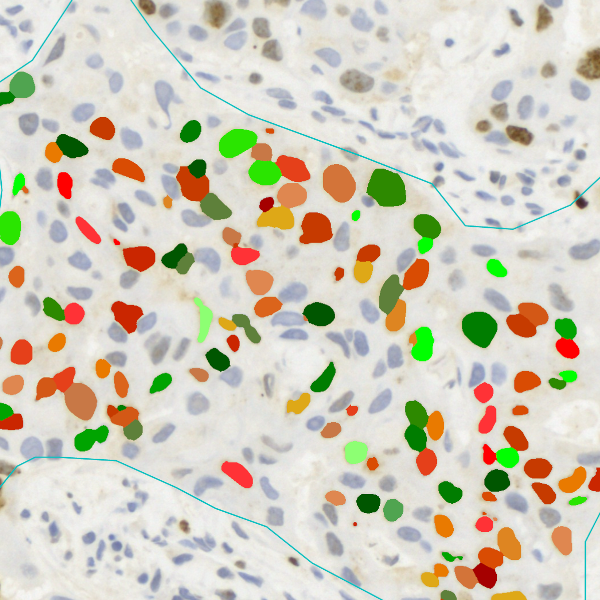
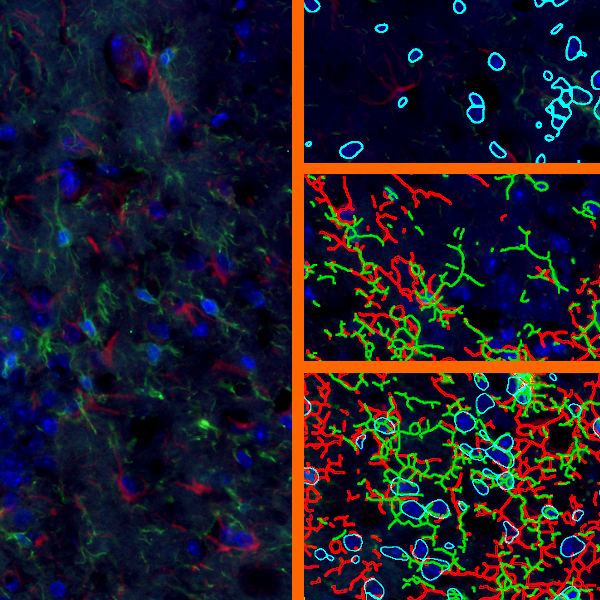
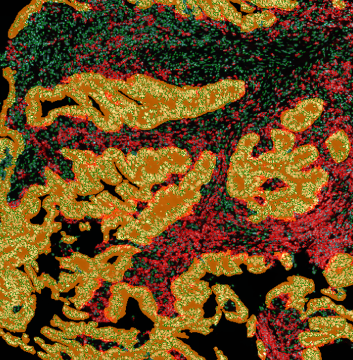
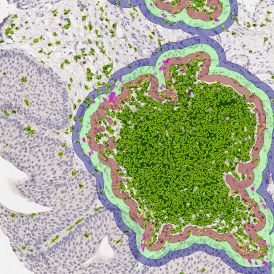
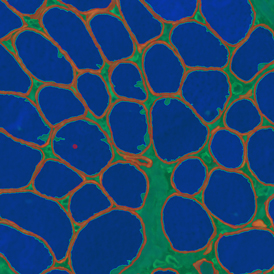
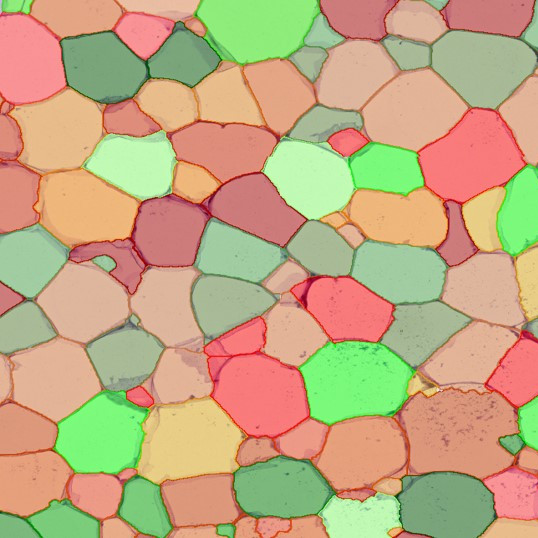
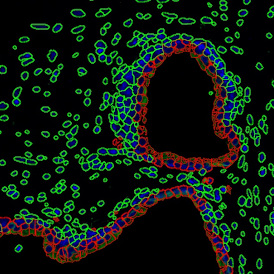
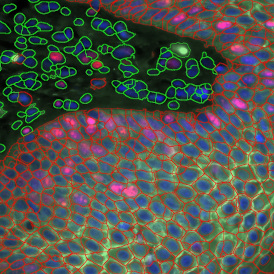
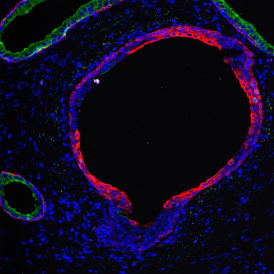
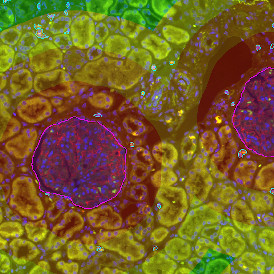
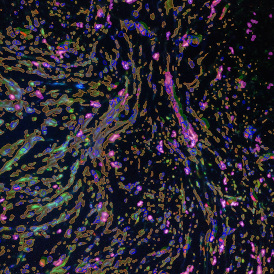
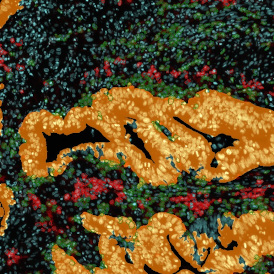
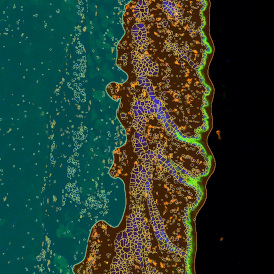
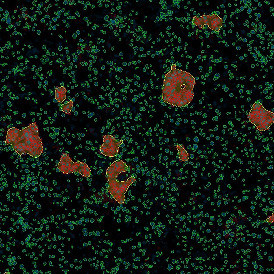
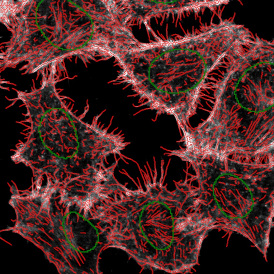
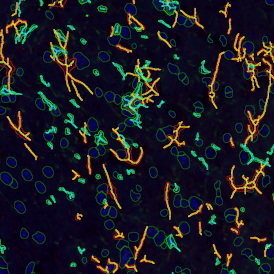
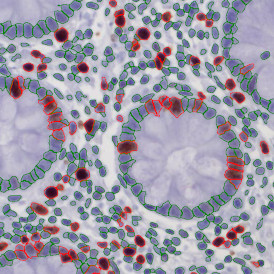
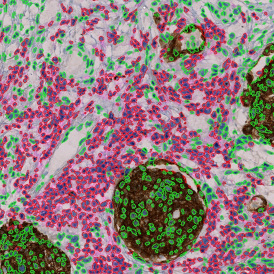
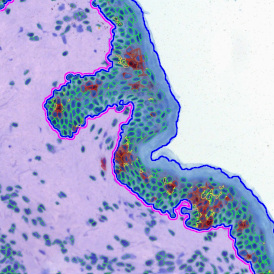
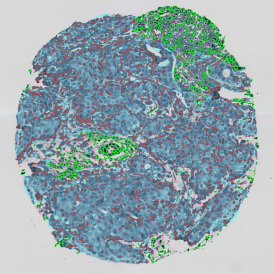
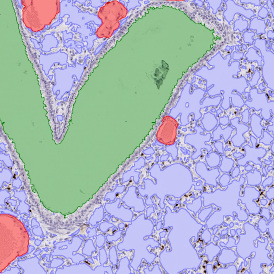
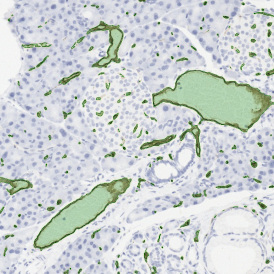
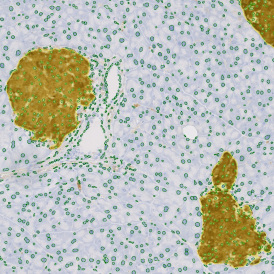
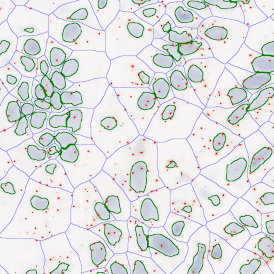
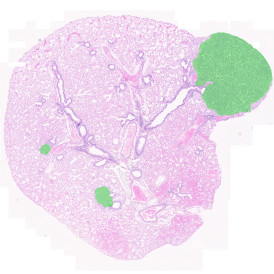
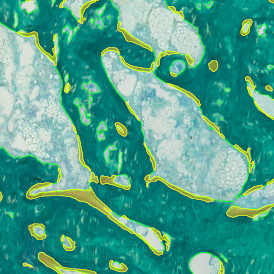
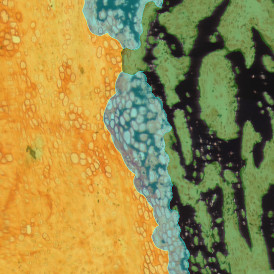
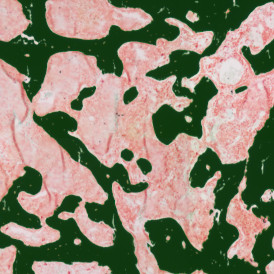
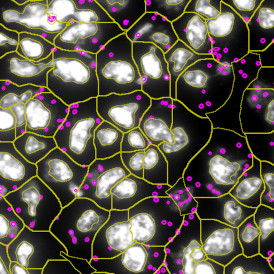
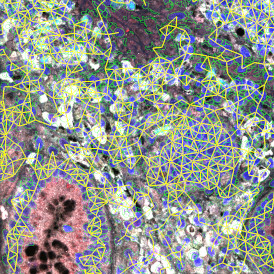
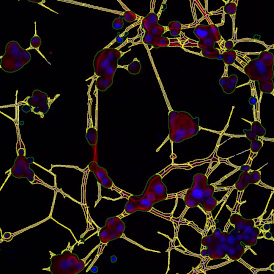
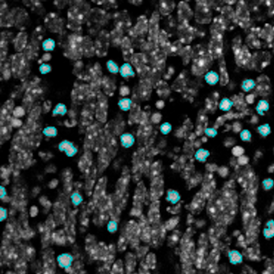
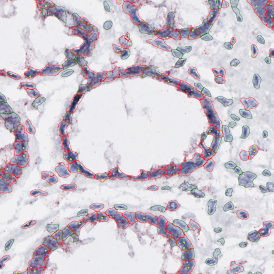
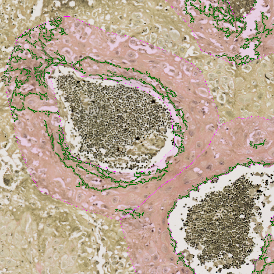
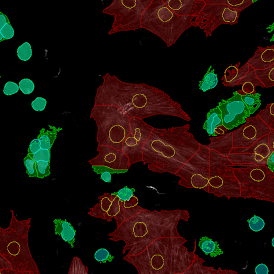
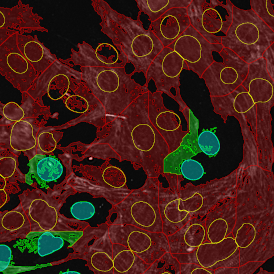
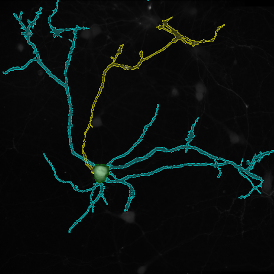
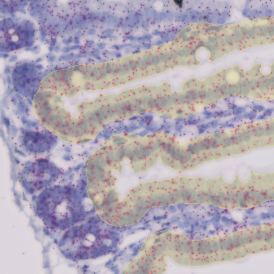
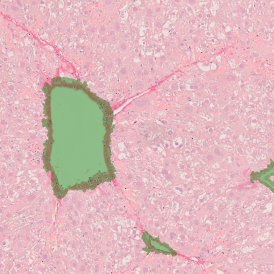
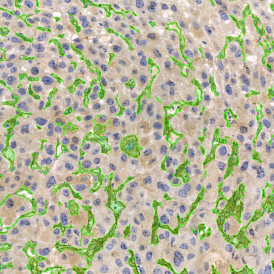
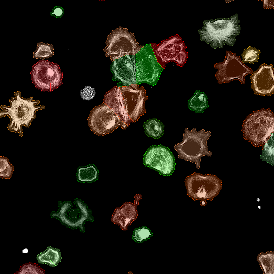
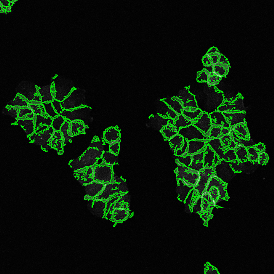
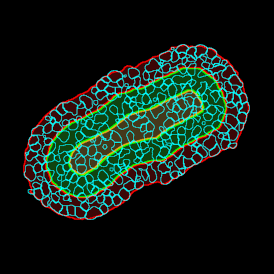
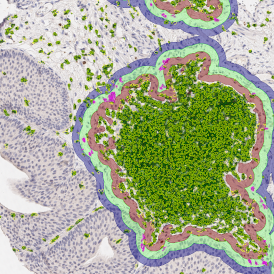
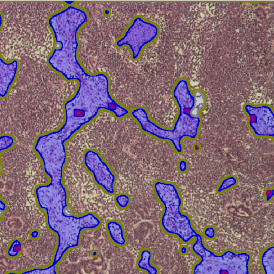
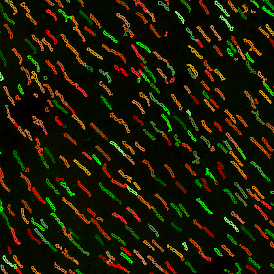
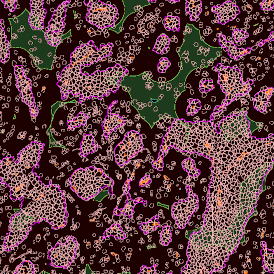
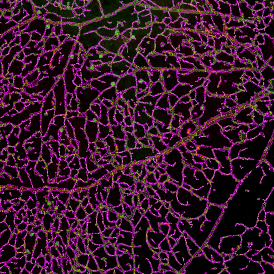
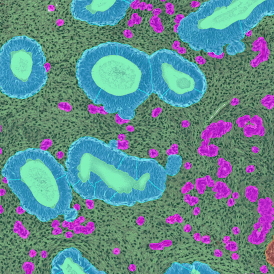
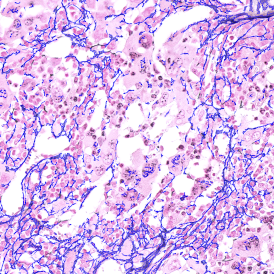
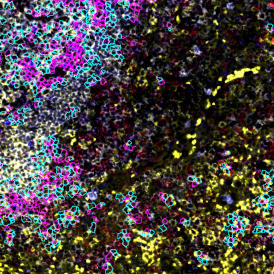
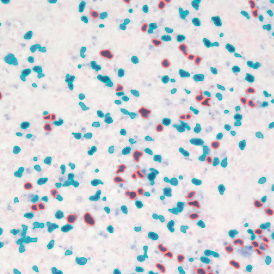
The IHC Meta.Cells APP combines the detection of IHC/HC stained metastructures (e.g. Langerhans islets) with single cell detection (segmentation of cells into nucleus, and/or perinuclear area and/or cytoplasm). Detected cells can be classified and visualized as being either within or outside of detected metastructures. Each detected area and cell compartment is measured for up to 20 intensity, statistic and morphometric parameters.
APP Category 2